This post has two motivations. First, it can serve as a future reference point if I need to mention tree types again, and of course it can be found via search engine by anybody who needs to look this stuff up. The second is my recent observation that some 'evolutionary' systematists have the tendency to call all phylogenetic trees cladograms, perhaps conflating ways of displaying evolutionary relationships with their dubious claim that cladists do not accept the existence of ancestors. I would like to take the opportunity to explain the different ways we can draw trees, what they mean, and what a cladogram really is.
Phylogenetic trees are simple non-cyclical graphs connecting terminals - often species - to show how the terminals are related. In species trees, the internal internodes are common ancestors, and the nodes where branches meet are speciation events. In gene trees, they would be ancestral alleles and mutation events, respectively.
Perhaps the oldest truly phylogenetic tree was drawn by Charles Darwin in his notebook, the famous "I think" diagram. But it was in that case an abstract model to help him visualise for himself common descent, not yet a concrete hypothesis about any specific organisms.
So we have a tree connecting terminals. I will further assume that the tree is outgroup-rooted, so that it has an explicit directionality: In the following examples, the arrow of time points from the left to the right. It could be different. All that follows would work just as well if the trees were turned by 90 degrees and the arrow of time pointed from bottom to top, or if the tree was circular as in the case of Darwin's sketch. What this post is about is simply what the branch lengths on the tree diagram mean, if anything.
Cladograms
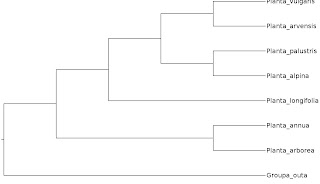
The least informative way of depicting a phylogenetic tree is as a cladogram. All that it shows is how the terminals are assumed to be related, nothing else. The branch lengths are meaningless and could be drawn with arbitrary length. But to show that this is the case, in practice people draw them either equal length or, as in the case of my example tree here, as all ending flush. If you are unsure if you are dealing with a cladogram, it might be useful to check if there is a scale bar on the diagram. If there isn't, it is probably a cladogram.
However, the author may still opt to put ticks onto the cladogram branches to illustrate where character changes took place. In that case, you will have the same information as provided by the phylogram (see below) but without meaningful branch lengths, so you are still dealing with a cladogram view of the tree.
If they are so uninformative, then why are cladograms used at all? As far as I can tell, in contemporary practice it has nothing to do with cladists' supposed dogmatic rejection of ancestors. Cladogram views are pragmatically used in phylogenetic publications when showing true branch lengths would lead to a very untidy and confusing looking tree or when there are no meaningful branch lengths. The latter is the case with consensus trees summarising a number of equally parsimonious trees or the results of bootstrapping or jackknifing. Each of the trees they are the consensus of had branch lengths, but the consensus tree itself shows only what relationships they agree on. It doesn't have well defined branch lengths on its own.
Phylograms
A phylogram is a phylogenetic tree whose branch lengths are proportional to how many character changes have been inferred along the branches. If the tree you are looking at has branches that do not end flush and a scale bar you are most likely dealing with a phylogram.
If the branch lengths are multiples of one, it is most parsimonious to assume that the tree is the result of a parsimony analysis. A length of one then means that one character change took place along the branch, two means two, and so on. If the branch lengths are tiny fractions of one, on the order of 0.004, the tree is most likely the result of a likelihood or Bayesian analysis. The length then means what fraction of the characters changed along the branch. I have no idea why parsimony and model-based phylogenies have such different conventions, but if you want to make them directly comparable you merely have to multiply all branch lengths in the likelihood tree with the number of characters in the original data matrix.
Note that in a phylogram view a zero length branch indicates that the common ancestor below that branch has been reconstructed to have the same characters as the descendant at the end of the branch. In my example tree, the common ancestor of Planta arvensis and Planta vulgaris would have been indistinguishable from Planta arvensis by the character set used in the analysis. An 'evolutionary' systematist is now free to pull a "this chimpanzee over there is my ancestor" and consider Planta arvensis to be the ancestor of Planta vulgaris. I do not think that makes sense, but the point is this is a question of approaches to classification. It is not a question of phylogenetic trees or cladistic analyses as such not allowing this interpretation.
Chronograms
A chronogram is a phylogenetic tree whose branch lengths are proportional to time. If the tree you are looking at is ultrametric, that is all branches end flush, and it has a full-length scale bar, you may be dealing with a chronogram. If the scale bar is in units of "Myr" or suchlike and starts with zero in the present you are definitely dealing with a chronogram.
Some chronograms may not be ultrametric because they contain extinct species, but the kind of fancy analysis that produces these kinds of trees is still rarely used, not least because many groups don't have decent fossils available anyway.
Phenograms
Another term that you may run into is phenogram, but this one is not about the meaning of branch lengths. Many systematists do not consider clustering by similarity to have a true phylogenetic logic whereas others disagree and consider it simply another tool in the phylogentic toolbox. The former accordingly use phenogram to differentiate the results of distance based, clustering, phenetic analyses from the phylogenetic trees resulting from what they consider to be actual phylogenetic analyses. Similarly, one would then call a group in a phenogram a cluster as opposed to clade, reserving the latter word for phylogenetic trees.
No plants were harmed in the making of this post. FigTree was used to produce the example trees.




Clear, succinct explanation. Thanks!
ReplyDelete