Last post we looked at what patrocladistics is and how it works. The example case was not a real group of organisms but contrived, but it was perhaps typical in that the dataset only included extant species.
In this post I want to explore what happens to the results of a patrocladistic analysis if we add all the ancestors. There are two reasons why this is of interest to me:
First, I believe that like many other ideas for the objective delimitation of paraphyletic taxa patrocladistics relies on not having intermediate ancestors in the dataset. Not all, perhaps, but many such approaches identify a long branch or gap in variation. The problem is that such a long branch or gap is merely an illusion based on the patchiness of the fossil record. In reality, evolution is gradual. And if somebody claims to have a good approach to classification it could be argued that it should be able to deal with the discovery of intermediate fossils.
Second, proponents of paraphyletic taxa often criticise phylogenetic systematists for supposedly ignoring ancestors, or for supposedly defining them out of existence. If patrocladistics does, as I suspected, rely on the absence of ancestors, that might at least be seen as a bit ironic.
So back to our artificial phylogeny. It contains two outgroup species and five ingroup species, two of the latter on a very long branch:
Using the patrocladistic approach with single-linkage clustering, I was able to produce a dendrogram that shows the two divergent species outside of the cluster of the other three ingroup species. A paraphyletic group on the phylogeny comes out as a cluster in the dendrogram, supposedly providing a justification for official taxonomic recognition:
Now assume an 'evolutionary' classification has become widely accepted that treats the species aberrans and anomalica in one genus and primitiva, communis and vulgaris in another. Imagine further that we are as lucky as palaeontologists have been with the transition between non-avian dinosaurs and birds. Every year sees another intermediate fossil published, and after a few years our phylogram looks like this:
Every letter is an intermediate ancestor. Note that I do not consider this to be a very realistic scenario. In most real life cases we would only have some of these letters. I am just saying that a general principle for classification should be able to deal with intermediate ancestors, especially if its proponents claim that supposedly not being able to do so is a major failure of the mainstream approach. And I am personally curious to see what happens if we repeat the patrocladistic analysis. First the new patristic distances:
Now the cladistic distances... aha. First interesting observation: If we have every intermediate 'species' according to the composite species concept, cladistic distances equal patristic distances. Every trait change on the branch has turned into a node. Because the patrocladistic distances are cladistic plus patristic, they are now just 2 x cladistic. Because that multiplication with two comes out in the wash distance-wise, we can as well proceed directly from the cladistic distance matrix to the clustering analysis.
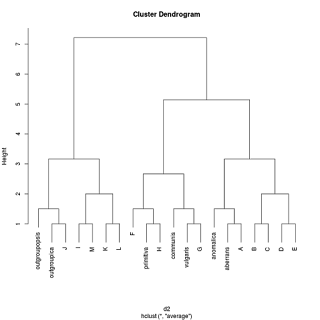
Using again the single-linkage clustering option in R's hclust function, the new dendrogram looks like this:
This was not quite what I expected; it was even worse! No clusters at all. But when we think about it, it is not surprising. Remember that single-linkage clustering always unites the two clusters that have the shortest distance between any two of their elements, so in a sense the shortest distance between their margins or outliers. But in this case, no matter how you cut the cake, there is never any distance larger than one. So everything immediately gets lumped into one flat cluster.
In a way I find this quite fitting. As mentioned above, evolution is gradual; clustering its results into paraphyletic taxa never made sense to me in the first place. And of course if we try to cluster by long branches then taking the historical, real life non-existence of long branches into account will make clustering impossible. One could now simply conclude that patrocladistics does indeed only work in the fortuitous absence of intermediate fossils and leave it at that.
But out of interest I tried a different clustering method provided by hclust, "average". Result:
Now we have a garbled version of the original phylogenetic tree back, except primitiva isn't in the right position and the ancestors are sister to their descendants, something that is anathema to many proponents of paraphyletic taxa.
Does that make sense? I think representatives of all schools of classification might actually agree that it doesn't. But the question is whether a patrocladistic analysis without ancestors makes any more sense. What is the theoretical background and justification? How do we interpret the results from a biological perspective?






I won't spoil again your last questions ;)
ReplyDeleteConcerning your other points, if I were to examine your UPGMA patrocladogram closer, I would say that you have indeed proven that primitiva-vulgaris-communis-F-H-G are a distinct evolutionary grade. From them arises a second grade made of B-C-D-E, which leads to a third evolutionary grade anomalica-aberrans-A.
In fact, I am surprised that patrocladistics works so well with so few species and so much ancestors included. I didn't expect it, and maybe I should revise my opinion that patrocladistics doesn't work with paleontological data. It seems it does.
You've just proven that single-linkage should be avoided.
Apparently this shows that every ancestor is a separate grade from its descendant. Aren't you usually against ancestors being treated as terminals? I guess that is only a problem in cladograms?
DeleteI am not sure to understand your first sentence, but if you mean that every species is a grade, then yes it's obvious. But since grades are nested into grades, the important thing to note is that on your patrocladogram there are 3 higher grades that match evolutionists' conceptualization of a natural classification.
DeleteThe only very wrong sentence of your post is your conclusion: "Does that make sense? I think representatives of all schools of classification might actually agree that it doesn't." The patrocladogram you show makes sense for evolutionists.
I don't understand where you see that ancestors are treated as terminals. You drew them right on the branches of your cladogram, thus transforming it into a true phylogenetic tree. Now you just have to draw circles on it to mark out the 3 genera I mentioned. Voilà! You have made your first evolutionary classification, congratulations!
Perhaps I was unclear. If a cladogram shows as sister two species you only interpret as ancestor and descendant (despite both existing contemporaneously) you go boo! hiss! cladists are [insert philosophical school here] because they don't recognise ancestors (and that despite the fact that a phylogram representation of the cladogram would pre-empt your complaint anyway).
DeleteIn a patrocladogram with average-clustering the actual, bona fide ancestor gets placed on a different branch with an actual length, and you have no problem.
Be that as it may, I can also draw other circles and make my first baraminological classification. Doesn't necessarily mean that it has any scientific value or merit though.
I am sorry you are not clearer (besides, using slang doesn't help me!).
DeleteAs I understand, you seem to think that evolutionists never draw cladograms where ancestors are represented as sisters to their descendants. This is wrong. The only difference is that we know that a cladogram is nothing but a tool, it doesn't represent the real phylogenetic tree. Similarly, a patrocladogram is nothing but a tool used to find grades. Being in the same cluster on a patrocladogram means being in the same grade, not being "sisters". Perhaps is it the tree-like picture that confuses you.
What seems unscientific to me is to pretend that something that exist (grades for example) can't be classified. At least, saying that they shouldn't be classified is dogmatic. You have proven that even if we include all the ancestors, we can still uncover the putative grades by using patrocladistics. If I were to write a post on a blog in order to convince people to use patrocladistics so as to determine and name the grades, I would not have done otherwise.
The only difference between cladogram and phylogram is the absence of branch lengths in the former. There are lots of things that 'exist' the moment we draw a red circle around them. Hey, my and my neighbour's cars are sitting next to each other! We need to recognise that!
DeleteIt is astonishing with what a tone of condescension you argue for what is a fringe position last relevant in the 1970ies.
You just refuse to recognize that besides the branching pattern there are other meaningful patterns in the phylogenetic trees, even after you proved yourself that such a pattern is consistently uncovered again after the inclusion of the ancestors, and hence is not an artifact due to missing data as you assumed.
DeleteI am sorry you are astonished but I don't understand what's wrong with my tone.
Are we looking at the same dendrograms? Without ancestors, we get clusters, with ancestors, we don't. Now you are using average clustering instead, and that seems to work more consistently. Fine, although interestingly it just recovers the original phylogeny despite a very long branch in the ingroup.
DeleteBut that brings us to the biological, theoretical or practical justification. If I cluster by whether species have yellow flowers I will also find a pattern in the phylogenetic tree, and I will also consistently uncover it when re-analysing: look, the yellow ones always go together! It's objective! Now: so what?
And that is before mentioning that clustering will always give you clusters even if, as in the case of having all the ancestors, there are no meaningful clusters to be had.
And it is not even mentioning the usual character selection issues. As K&S themselves pointed out, the results of patrocladistics will be very different for different data sets. They seem happy to use DNA sequence data; you don't. They talk of apomorphies having pre-selected as phylogenetically important; you talk of 'adaptive zones', but that means you have to pre-select adaptively important instead of phylogenetically important characters. That's where the objectivity comes crashing down once more.
"it just recovers the original phylogeny" Hu no, look again. There are the three grades I have described. It's not the original phylogeny. With or without ancestors you get the same clusters using UPGMA.
Delete"If I cluster by whether species have yellow flowers I will also find a pattern in the phylogenetic tree" If yellow is not an apomorphy your yellow flowers won't be clustered together because of their yellowness. You are confusing phenetic and patristic distances.
"you have to pre-select adaptively important instead of phylogenetically important characters." Hu no, where have I said such a thing? I already said that all available characters should be pooled together.
You are imagining things, stop attack straw mans.
Ah, sorry, I just did the average clustering again, and indeed it clusters primitiva with communis and vulgaris. So that at least is consistent. Clusters still don't make any more sense than phenetic ones; less actually.
DeleteI feel less like attacking strawmen than like trying to nail a pudding to the wall. Your justification is about adaptive zones, and you have explicitly rejected the use of molecular data. I assume further that you will not seriously argue that a long branch based on apomorphic but adaptation-irrelevant characters like opposite vs. alternate leaves, toothed vs. entire leaf margin, scabrous vs. glabrous upper leaf surface, even-pinnate vs. odd-pinnate leaf, and yellow vs. white flower separates two temperate zone forest trees adapted to calcareous soils into different adaptive zones?
How then is your total (but not really, only total morphological) evidence approach supposed to reflect adaptive zones?
This would be obviously circular to tell aprioristically that such and such adaptive zones exist, that such and such characters are those adapted to these zones, and then to draw a patrocladogram with only those characters and say "Look! I have proven that those characters are adaptations to particular adaptive zones!". Are evolutionary systematists really supposed to be such stupid people?
DeleteAll phenotypic characters should be used without apriori. Adaptive zones aren't defined before the analysis, they are discovered by the analysis. Cladistics proceeds likewise, synapomorphies aren't defined as such before the analysis!
Besides, there are many reasons why apparently non-adaptive characters could be correlated to adaptive zones. For example, they can be adaptive after all, even a weak selection pressure is efficient over long times. Or, they can be truly non-adaptive but result from a genetic drift that happened during the transition between two adaptive zones, so they are still useful landmarks indicating that a transition had occured.
There are some significant differences between the two situations. Cladists ask what characters are indicative of relatedness, and it is obvious that we can only answer that question once we have the phylogeny. In contrast, to the degree that people could ever agree on what adaptive zones and niches are and whether those concepts are actually useful it should be possible to define them in complete ignorance of phylogenetic relationships, even in the absence of common descent.
DeleteWhich leads us to the second point, which is again that there is not even so much as a tenuous relationship between branch length and adaptive zones. The cladist's logic works as follows:
1. I want to know which characters are indicative of relationships.
2. I infer a phylogeny.
3. Heritable traits that show up on the phylogeny as having evolved in a common ancestor are necessarily indicative of relatedness of the descendants of that common ancestor (while allowing for some secondary transformations on top of that further down the line).
One may argue about this approach but the last step follows from the previous ones. Your logic is either:
1. I want to recognise paraphyletic taxa if they are in different adaptive zones.
2. I use as many non-molecular characters as I can get, a few of which "could be related to" adaptive zones, to make clusters.
3. ????
Or, alternatively:
1. I use as many non-molecular characters as I can get, a few of which "could be related to" adaptive zones, to make clusters.
2. ????
3. The clusters are in different adaptive zones!
Not sure which it is really, but either way some crucial step is missing. At least K&S did not claim any ecological rationale. Their focus on what looks like anagenetic divergence appears misguided to me because there must have been intermediate ancestors and potentially extinct side branches along the whole branch, consequently the gap they want to recognise is just an illusion, and consequently there are no meaningful clusters. But as they don't say they care about adaptive zones they don't need to care about the adaptive value of the trait changes along the branch.
Once more, *I* *do* care about adaptive zones. I am happy to recognise the polyphyletic group "arid zone ephemeral", but mixing that up with paraphyletic taxa seems like a category error.
"consequently the gap they want to recognise is just an illusion" You have consistently proven the contrary. Three grades are recognized even after the inclusion of all ancestors.
Delete"Once more, *I* *do* care about adaptive zones." You confuses adaptive zones with ecological niches.
The evolutionary approach is not the caricature you describe.
1. I want to recognise paraphyletic taxa if they are in different adaptive zones.
2. Transition between adaptive zones implies a shift in phenotypic rate of evolution.
3. A shift in phenotypic rate of evolution implies some kinds of imbalance in the shape of the phylogenetic trees.
4. I map as many phenotypic characters as I can get on the phylogenetic tree, and I apply a clustering algorithm that can account for the kind of imbalance I am looking for.
5. The grades found by the clustering algorithm are assumed to correspond putative adaptive zones.
6. I can test this assumption against paleontological or ecological data, or by adding more data.
7. The resulting classification is more natural because it accounts for the whole process of evolution and not for the topological pattern alone.
The refusal to consider paraphyletic taxa only stems only from the rejection of points 1 and/or 7, i.e. respectively dogmatism or structuralist philosophy.
Three grades are recognized even after the inclusion of all ancestors.
DeleteI repeat: clustering will always give you clusters even if there are no meaningful clusters to be had. Which is why I was so surprised that single-linkage actually didn't. Try it: make some random numbers, use them as distances, and cluster. You will get clusters.
In this case, single is the method that gets it right: There really were no distinct clusters along the tree of life, everything is seamlessly connected. Average-clustering just gives you an illusion. I have seen people make that mistake before, only in the context of vegetation analysis.
1. No doubt.
2. Why? How? That is news to me. It only necessarily involves a shift in the very few traits that are adaptation relevant.
3. Not necessarily, depends on speciation and extinction rates (which could just as well go up as down when shifting to a new peak), pure chance, and more besides.
4. I would find it more productive to do a BAMM analysis or something comparable; that is how evolutionary biologists work these days.
5. That assumption is completely unfounded, see above.
6. Adding more non-adaptive traits will not tell you more about adaptation.
7. The resulting classification is inconsistent with the approaches everybody uses at lower levels (tokogeny & ontogeny), internally inconsistent, and misleading to the end user, it has not use case beyond your own movement's desire to have paraphyletic taxa, and due to the problems with 2-6 it wouldn't even reflect what you want it to reflect.
Also, this is not how the "evolutionary" approach works. Nearly every "evolutionary" systematist has a completely idiosyncratic approach.
"clustering will always give you clusters" Bootstrap, and meaningless clusters will vanish in a polytomy. Try it with random numbers vs non-random numbers. No illusion in this. The result doesn't satisfy you so you discard it. Too easy.
Delete2. Even if the shift implies very few traits, the change is going to be quick (directional selection + genetic drift), while the traits remained conserved in the former zone (stabilizing selection + Hardy-Weinberg equilibrium).
3. Speciation and extinction rates depends on point 2, especially the ratio between cladogenetic speciation and anagenetic speciation.
4. Yes, through necessary data are not always available. That's why patrocladistics should be considered an heuristic. Just like parsimony-based inference of phylogeny is an heuristic to model-based inference.
5. No, it is consistent.
6. Will test the consistency of the clustering. The fact that the approach is falsifiable is a strength, not a weakness.
7. Evolutionary systematics is consistent with tokogeny and ontogeny, a hen does not become another hen after laying an egg. I don't see any proof of internal inconsistency. End users are disappointed that species grouped in the same genus are not similar to each other but to species grouped in other genera. As for use case, this classification is more stable that cladistic one and hence more useful to end users (see Ruggiero's choice).
Results between evolutionary systematists are consistent and methodology is converging. I could also say that with parsimony, ML or BI, nearly every phylogeneticist has an idiosyncratic approach...
I guess the difference here is that you select the clustering method to give you clusters, and then you are happy; while I look at the tree, see that there are no clusters to be had because evolution is gradual, and further see no logic in clustering or in using patristic + cladistic distances in the first place. To me it reads like "steal socks, ????, profit!"
DeleteSometimes I wonder if you are simply using random words that sound profound. Are you implying that there is no genetic drift with stabilising selection, or no Hardy-Weinberg in directional selection? You are aware that at any point only a minuscule part of the genome could ever be under directional selection, right? Are you implying that a shift to a new adaptive solution will always be accompanied by a higher diversification rate?
7: Point being, end users are somehow not disappointed that caterpillars of different species are more similar to each other than to the imagines of their own species. The task of systematists is to find out what indicates relatedness.
How can a classification be stable if it allows scientific progress? Apart from that, even your clustering would have to change if you discover a great extinct diversification instead of the long thin branch you assumed before.
You are conflating tree inference methods (which in practice nearly always produce the same results anyway) with the principle: "all supraspecific taxa should be monophyletic". Once we have a phylogeny, phylogenetic systematists know what groups are not acceptable. At that point your problems only start.
And just read through that Annals MO Bot Gard special issue edited by Hoerandl and Stuessy. Is anybody except Zander using his "Bayesian solution"? Is anybody except George arguing for accepting a genus even if it turns out to be polyphyletic? Is anybody except Stuessy using patrocladistics? Is even just one person in that issue making the same argument as you? Taken at face value, everybody has their personal concerns, but the only thing they agree on is that paraphyletic taxa should be accepted.
"there are no clusters to be had because evolution is gradual" Non sequitur.
Delete"Sometimes I wonder if you are simply using random words that sound profound." Isn't it what cladists always do?
"a shift to a new adaptive solution will always be accompanied by a higher diversification rate?" Always a higher phenotypic rate. If diversification = cladogenesis only, then no this won't always happen.
"end users are somehow not disappointed that caterpillars of different species are more similar to each other than to the imagines of their own species." Straw man.
"The task of systematists is to find out what indicates relatedness." Circular reasoning. Relatedness is only a part of the job.
"even your clustering would have to change" Yes, of course. More stable doesn't mean immutable.
"You are conflating" No, I was making a comparison.
"Once we have a phylogeny, phylogenetic systematists know what groups are not acceptable." And thus they reject natural groups.
"At that point your problems only start." I understand the appeal of cladism is that it needs less work.
We are now going into circles again. To make things clearer, it seems to me you could argue against paraphyly only in three ways.
1. Evolutionary patterns are explainable by Brownian motion only.
2. The methodology used cannot accurately uncover adaptive zones.
3. The classification should not reflect adaptive zones.
As far as I know, point 1 is wrong, it's a fact. Concerning point 2, if the methodology is not good enough then this just means that it should be replaced by a better one. As long as point 1 is rejected, such a methodology exist and should be discovered. Point 3 is at worst dogmatism, at least bad philosophy.
It is not clear to me how making a rule that classifications should reflect adaptive zones would be any less dogmatic than making a rule that classifications should reflect relatedness.
DeleteWe are now going into circles again.
That, at least, we can agree on. Hallelujah! A meeting of the minds has been achieved. Going to get some work done now.
So, this is maybe tangential to your real interest (systems of classification and naming, which, uh, I stay out of), but I'd point out that if you tried tracing actual character evolution (say, morphological), you might find that its very hard to get much phylogenetic resolution when you have sampled ancestors. And, if you have ancestors that can persist through branching events (i.e. budding, which you don't seem to allow for in your example; it all looks like bifurcation to me), that raises even more problems for trying to resolve relationships among units:
ReplyDeletehttp://www.ncbi.nlm.nih.gov/pubmed/23638034
The assumption here is that a new species is defined by having a new character, whether that makes biological sense or not. So under this assumption, budding may happen, whether that term is actually useful beyond motivated reasoning or not. And as long as there is no homoplasy (which is really the result of our ignorance of true homology) we would expect to get one most parsimonious tree that shows any 'ancestors' sitting on a zero length branch in phylogram view, just like my tree with the letters along the branches. Another little contrived example, this time with lots of budding, is here.
DeleteThe reason I did not put any zero length branches onto the original example phylogeny in this case is because I wanted to keep ancestors out of the first analysis and then only add them in the second. They are two extreme case scenarios, if you will.
An excellent paper David! These are the kind of examples I have in mind when I try to explain why the internodal species concept is misleading, or the idea that a complete cladification is possible. I must definitely cite you next time.
Delete