Because I am fed up with figuring it out anew every time I need to use the Incongruence Length Difference (ILD) test (Farris et al., 1994) in TNT, I will post it once and for all here:
Download TNT and the script "ildtnt.run" from PhyloWiki. In the script, you may have to replace all instances of "numreps" with "num_reps" to make it functional. I at least get the error "numreps is a reserved expression", suggesting that the programmer should not have used that as a variable name.
Open TNT, increase memory, and set data to DNA and treating gaps as missing data. Then load your data matrix, which should of course be in TNT format:
mxram 200 ;
nstates DNA ;
nstates NOGAPS ;
proc (your_alignment_file_name) ;
Look up how many characters your first partition has, then run the test with:
run ildtnt.run (length_of_first_partition) (replicates) ;
There is an alternative script for doing the test called Ild.run, but I have so far failed to set the number of user variables high enough to accommodate my datasets. They seem to be limited to 1,000?
Perhaps this guide will also be useful to somebody besides me.
Reference
Farris JS, Källersjö M, Kluge AG, Bult C, 1994. Testing significance of incongruence. Cladistics 10: 315-319.
Showing posts with label how to. Show all posts
Showing posts with label how to. Show all posts
Thursday, September 26, 2019
Friday, April 20, 2018
Time-calibrated or at least ultrametric trees with the R package ape: an overview
I had reason today to look into time-calibrating phylogenetic trees again, specifically trees that are so large that Bayesian approaches are not computationally feasible. It turns out that there are more options in the R package APE than I had previously been aware of - but unfortunately they are not all equally useful in everyday phylogenetics.
In all cases we first need a phylogram that we want to time-calibrate or at least make ultrametric to use in downstream analyses that require ultrametricity. As we assume that our phylogeny is very large it may for example have been inferred by RAxML, and the branch lengths are proportional to the probability of character changes having happened along them. For present purposes I have used a smaller tree (actually a clade cut out of a larger tree I had floating around), so that I could do the calibrations quickly and so that the figures of this post look nice. My example phylogram has this shape:
We fire up R, load the ape package, and import our phylogeny with read.tree() or read.nexus(), depending on whether it is in Newick or Nexus format, e.g.
Penalised Likelihood
I have previously done a longer, dedicated post on this method. I did not, however, go into the various models and options then, so let's cover the basics here.
Penalised Likelihood (PL) is, I think, the most sophisticated approach available in APE, allowing the comparison of likelihood scores between different models. It is also the most flexible. It is possible to set multiple calibration points, as discussed in the linked earlier post, but here we simply set the root age to 50 million years:
Let's start with correlated:
Next, discrete. The command is the same as above except for the text in the model parameter. The branch length distribution and likelihood score turned out to be very close to those for the correlated model:
Finally, relaxed. Very different branch length distribution and a by far worse likelihood score compared to the other two:
I have only considered testing a strict clock model with chronos for the first time today. It turns out that you get it by running it as a special case of the discrete model, which by default is set to assume ten rate categories. You simply set the number of categories to one:
The problem with PL is that is seems to be a bit touchy. Even today we had several cases of an inexplicable error message, and several cases of the analysis being unable to find a reasonable starting solution. We finally found that it helped to vastly increase the root age (we had played around with 15, assuming that it doesn't matter, and it worked when we set it to a more realistic three digit number). It is possible that our true problem was short terminal branches.
PL is also the slowest of the methods presented here. I would use it for trees that are too large for Bayesian time calibration but where I need an actual chronogram with a meaningful time axis and want to do model comparison. If I just want an ultrametric tree the following three methods would be faster and simpler alternatives. That being said, so far I had no use case for them.
A superseded but fast alternative: chronopl()
This really came as a surprise as I believed that the function chronopl() had been removed from ape. I thought I had tried to find it in vain a few years ago, but I saw it in the ape documentation today (albeit with the comment "the new function chronos replaces the present one which is no more maintained") and was then able to use it in my current R installation. I must have confused it with a different function.
chronopl() does not provide a likelihood score as far as I can see, but it seems to be very fast. I quickly ran it with default parameters and lambda = 1, again setting root age to 50:
Various parameters can be changed, but as implied above, if I want to do careful model comparison I would use chronos() anyway.
Mean Path Lengths
The chronoMPL() method time-calibrates the phylogeny with what is called a mean path lengths method. The documentation makes clear that multiple calibration points cannot be used; the idea is to make an ultrametric tree, pick one lineage split for which one has a credible date, and then scale the whole tree so that the split has the right age. Command is simply:
Most of the branch length distribution fits the results for the favoured model in the analysis with chronos(), see above. That's actually great, because chronoMPL() is so much faster! But you will notice some wonky lines in particular in the top right and bottom right corners of this tree graph. Those are negative branch lengths. Did somebody throw the ancestral species into a time machine and set them free a bit before they actually evolved?
Some googling suggests that this happens if the phylogram is very unclocklike, which, unfortunately, is often the case in real life. That limits rather sharply what mean path lengths can be used for.
The compute.brtime() function
Another function that I have now tried out is compute.brtime(). It can do two rather different things.
The first is to transform a tree according to what I understand has be a full set of branching times for all splits in the tree. The use case for that seems to be if you have a tree figure and a table of divergence times in a published paper and want to copy that chronogram for a follow-up analysis, but the authors cannot or won't send it to you. So you manually type out the tree, manually type out a vector of divergence times (knowing which node number is which in the R phylo format!), and then you use this function to get the right branch length distribution. May happen, but presumably not a daily occurrence. What we usually have is a tree for which we want the analysis to infer biologically realistic divergence times that we don't know yet.
The second thing the function can do is to infer an ultrametric tree without any calibration points at all but under the coalescent model. The command is then as follows.
Note that this is more on the lines of "one possible solution under the coalescent model" instead of "the optimal solution under this here clock model", so that every run will produce a slightly different ultrametric tree. I ran it a few times, and one aspect that did not change was the clustering of nearly all splits close to the present, which I (and PL, see above) would consider biologically unrealistic. Still, we have an ultrametric tree in case we need one in a hurry.
It is well possible that I have still missed other options in APE, but these are the ones I have tried out so far.
Something completely different: non-ultrametric chronograms
Finally, I should mention that there are methods to produce very different time-calibrated trees in palaeontology. The chronograms discussed in this post are all inferred under the assumption that we are dealing with extant lineages, so all branches on the chronogram end flush in the present, and consequently a chronogram is an ultrametric tree. And usually the data that went into inferring the topology was DNA sequence data or similar.
Palaeontologists, however, deal with chronograms where many or all branches end in the past because a lineage went extinct, making their chronograms non-ultrametric and look like phylograms. And usually the data that went into inferring the tree topology was morphological. This is a whole different world for me, and I can only refer to posts like this one and this one which discuss an R package called paleotree.
There also seems to be a function in newer APE versions called node.date() which is introduced with the following justification:
In all cases we first need a phylogram that we want to time-calibrate or at least make ultrametric to use in downstream analyses that require ultrametricity. As we assume that our phylogeny is very large it may for example have been inferred by RAxML, and the branch lengths are proportional to the probability of character changes having happened along them. For present purposes I have used a smaller tree (actually a clade cut out of a larger tree I had floating around), so that I could do the calibrations quickly and so that the figures of this post look nice. My example phylogram has this shape:
We fire up R, load the ape package, and import our phylogeny with read.tree() or read.nexus(), depending on whether it is in Newick or Nexus format, e.g.
mytree <- read.tree("treefilename.tre")Now to the various methods.
Penalised Likelihood
I have previously done a longer, dedicated post on this method. I did not, however, go into the various models and options then, so let's cover the basics here.
Penalised Likelihood (PL) is, I think, the most sophisticated approach available in APE, allowing the comparison of likelihood scores between different models. It is also the most flexible. It is possible to set multiple calibration points, as discussed in the linked earlier post, but here we simply set the root age to 50 million years:
mycalibration <- makeChronosCalib(mytree, node="root", age.max=50)We have three different clock models at our disposal, correlated, discrete, and relaxed. Correlated means that adjacent parts of the phylogeny are not allowed to evolve at rates that are very different. Discrete models different parts of the tree as evolving at different rates. As I understand it, relaxed allows the rates to vary most freely. Another important factor that can be adjusted is the smoothing parameter lambda; I usually run all three clock models at lambdas of 1 and 10 and pick the one with the best likelihood score. For present purposes I will restrict myself to lambda = 1.
Let's start with correlated:
mytimetree <- chronos(mytree, lambda = 1, model = "correlated", calibration = mycalibration, control = chronos.control() )When plotted, the chronogram looks as follows.
Next, discrete. The command is the same as above except for the text in the model parameter. The branch length distribution and likelihood score turned out to be very close to those for the correlated model:
Finally, relaxed. Very different branch length distribution and a by far worse likelihood score compared to the other two:
I have only considered testing a strict clock model with chronos for the first time today. It turns out that you get it by running it as a special case of the discrete model, which by default is set to assume ten rate categories. You simply set the number of categories to one:
mytimetree <- chronos(mytree, lambda = 1, model = "discrete", calibration = mycalibration, control = chronos.control(nb.rate.cat=1) )In my example case this looks rather similar to the results from correlated model and discrete with ten categories:
The problem with PL is that is seems to be a bit touchy. Even today we had several cases of an inexplicable error message, and several cases of the analysis being unable to find a reasonable starting solution. We finally found that it helped to vastly increase the root age (we had played around with 15, assuming that it doesn't matter, and it worked when we set it to a more realistic three digit number). It is possible that our true problem was short terminal branches.
PL is also the slowest of the methods presented here. I would use it for trees that are too large for Bayesian time calibration but where I need an actual chronogram with a meaningful time axis and want to do model comparison. If I just want an ultrametric tree the following three methods would be faster and simpler alternatives. That being said, so far I had no use case for them.
A superseded but fast alternative: chronopl()
This really came as a surprise as I believed that the function chronopl() had been removed from ape. I thought I had tried to find it in vain a few years ago, but I saw it in the ape documentation today (albeit with the comment "the new function chronos replaces the present one which is no more maintained") and was then able to use it in my current R installation. I must have confused it with a different function.
chronopl() does not provide a likelihood score as far as I can see, but it seems to be very fast. I quickly ran it with default parameters and lambda = 1, again setting root age to 50:
mytimetree <- chronopl(mytree, lambda = 1, age.min = 50, age.max = NULL, node = "root")The result looks very similar to what chronos() produced with the (low likelihood) relaxed model:
Various parameters can be changed, but as implied above, if I want to do careful model comparison I would use chronos() anyway.
Mean Path Lengths
The chronoMPL() method time-calibrates the phylogeny with what is called a mean path lengths method. The documentation makes clear that multiple calibration points cannot be used; the idea is to make an ultrametric tree, pick one lineage split for which one has a credible date, and then scale the whole tree so that the split has the right age. Command is simply:
mytimetree <- chronoMPL(mytree)The problem is, the resulting chronogram often looks like this:
Most of the branch length distribution fits the results for the favoured model in the analysis with chronos(), see above. That's actually great, because chronoMPL() is so much faster! But you will notice some wonky lines in particular in the top right and bottom right corners of this tree graph. Those are negative branch lengths. Did somebody throw the ancestral species into a time machine and set them free a bit before they actually evolved?
Some googling suggests that this happens if the phylogram is very unclocklike, which, unfortunately, is often the case in real life. That limits rather sharply what mean path lengths can be used for.
The compute.brtime() function
Another function that I have now tried out is compute.brtime(). It can do two rather different things.
The first is to transform a tree according to what I understand has be a full set of branching times for all splits in the tree. The use case for that seems to be if you have a tree figure and a table of divergence times in a published paper and want to copy that chronogram for a follow-up analysis, but the authors cannot or won't send it to you. So you manually type out the tree, manually type out a vector of divergence times (knowing which node number is which in the R phylo format!), and then you use this function to get the right branch length distribution. May happen, but presumably not a daily occurrence. What we usually have is a tree for which we want the analysis to infer biologically realistic divergence times that we don't know yet.
The second thing the function can do is to infer an ultrametric tree without any calibration points at all but under the coalescent model. The command is then as follows.
mytimetree <- compute.brtime(mytree, method="coalescent", force.positive=TRUE)It seems that the problem of ending up with negative branch lengths was, in this case, recognised and solved simply by giving the user the option to tell the function PLEASE DON'T. I assume they are collapsed to zero length (?). My result looked like this:
Note that this is more on the lines of "one possible solution under the coalescent model" instead of "the optimal solution under this here clock model", so that every run will produce a slightly different ultrametric tree. I ran it a few times, and one aspect that did not change was the clustering of nearly all splits close to the present, which I (and PL, see above) would consider biologically unrealistic. Still, we have an ultrametric tree in case we need one in a hurry.
It is well possible that I have still missed other options in APE, but these are the ones I have tried out so far.
Something completely different: non-ultrametric chronograms
Finally, I should mention that there are methods to produce very different time-calibrated trees in palaeontology. The chronograms discussed in this post are all inferred under the assumption that we are dealing with extant lineages, so all branches on the chronogram end flush in the present, and consequently a chronogram is an ultrametric tree. And usually the data that went into inferring the topology was DNA sequence data or similar.
Palaeontologists, however, deal with chronograms where many or all branches end in the past because a lineage went extinct, making their chronograms non-ultrametric and look like phylograms. And usually the data that went into inferring the tree topology was morphological. This is a whole different world for me, and I can only refer to posts like this one and this one which discuss an R package called paleotree.
There also seems to be a function in newer APE versions called node.date() which is introduced with the following justification:
Our software, node.dating, uses a maximum likelihood approach to perform divergence-time analysis. node.dating is written in R v3.30 and is a recent addition to the R package ape v4.0 (Paradis et al., 2004). Previously, ape had the capability to estimate the dates of internal nodes via the chronos function; however, chronos requires ultrametric trees and is thus unable to incorporate information from tips that are sampled at different points in time.This suggests that the point is the same, to allow chronograms with extinct lineages, but in this case aimed more at molecular data. Their example case are virus sequence data.
Wednesday, March 21, 2018
Bioregionalisation part 6: Modularity Analysis with the R package rnetcarto
Today's final post in the bioregionalisation series deals with how to do a network or Modularity Analysis in R. There are two main steps here. First, because we are going to assume, as in the previous post, that we have point distribution data in decimal coordinates, we will turn them into a bipartite network of species and grid cells.
We start by defining a cell size. Again, our data are decimal coordinates, and subsequently we will use one degree cells.
We make a list of all species and a list of all cells that occur in our dataset, naming the cells after their centres in the format "126.5:-46.5". I assume here that we have the data matrix called 'mydata' from the previous post, with the columns species, lat and long.
Once the analysis is done, we may first wonder how many modules, which we will subsequently interpret as bioregions, the analysis has produced.
As an example I have done an analysis with all Australian (and some New Guinean) lycopods, the dataset I mentioned in the previous post. It plots as follows.
There are, of course, a few issues here. The analysis produced six modules, but three of them, the green, orange and light blue ones, consist of only two, one and one cells, respectively, and they seem biologically unrealistic. They may be artifacts of not having cleaned the data as well as I would for an actual study, or represent some kind of edge effect. The remaining three modules are clearly more meaningful. Although they contain some outlier cells, we can start to interpret them as potentially representing tropical (red), temperate (yellow), and subalpine/alpine (dark blue) assemblies of species, respectively.
Despite the less than perfect results I hope the example shows how easy it is to do such a Modularity Analysis, and if due diligence is done to the spatial data, as we would do in an actual study, I would also expect the results to become cleaner.
We start by defining a cell size. Again, our data are decimal coordinates, and subsequently we will use one degree cells.
cellsize <- 1Note that this may not be the ideal approach for publication. The width of one degree cells decreases towards the poles, and in spatial analyses equal area grid cells are often preferred because they are more comparable. If we want equal area cells we first need to project our data into meters and then use a cellsize in meters (e.g. 100,000 for 100 x 100 km). There are R functions for such spatial projection, but we will simply use one degree cells here.
We make a list of all species and a list of all cells that occur in our dataset, naming the cells after their centres in the format "126.5:-46.5". I assume here that we have the data matrix called 'mydata' from the previous post, with the columns species, lat and long.
allspecies <- unique(mydata$species)We create a matrix of species and cells filled with all zeroes, which means that the species does not occur in the relevant cell. Then we loop through all records to set a species as present in a cell if the coordinates of at least one of its records indicate such presence.
longrounded <- floor(mydata$long / cellsize) * cellsize + cellsize/2
latrounded <- floor(mydata$lat / cellsize) * cellsize + cellsize/2
cellcentre <- paste(longrounded,latrounded, sep=":")
allcells <- unique(cellcentre)
mynetw <- matrix(0, length(allcells), length(allspecies))It is also crucial to name the rows and columns of the network so that we can interpret the results of the Modularity Analysis.
for (i in 1:length(mydata[,1]))
{
mynetw[ match(cellcentre[i],allcells) , match(mydata$species[i], allspecies) ] <- 1
}
rownames(mynetw) = allcellsNow we come to the actual Modularity Analysis. We need to have the R library rnetcarto installed and load it.
colnames(mynetw) = allspecies
library(rnetcarto)The command to start the analysis is simply:
mymodules <- netcarto(mynetw, bipartite=TRUE)This may take a bit of time, but after talking to colleagues who have got experience with other software it seems it is actually reasonably fast - for a Modularity Analysis.
Once the analysis is done, we may first wonder how many modules, which we will subsequently interpret as bioregions, the analysis has produced.
length(unique(mymodules[[1]]$module))For publication we obviously want a decent map, but that is beyond the scope of this post. What follows is merely a very quick and dirty way of plotting the results to see what they look like, but of course the resulting coordinates and module numbers can also be used for fancier plotting. We split the latitudes and longitudes back out of the cell names, define a vector of colours to use for mapping (here thirteen; if you have more modules you will of course need a longer vector), and then we simply plot the cells like some kind of scatter plot.
allcells2 <- strsplit( as.character( mymodules[[1]]$name ), ":" )
allcells_x <- as.numeric(unlist(allcells2)[c(1:(length(allcells)))*2-1])There we are. Modularity analysis with the R library rnetcarto is quite easy, the main problem was building the network.
allcells_y <- as.numeric(unlist(allcells2)[c(1:(length(allcells)))*2])
mycolors <- c("green", "red", "yellow", "blue", "orange", "cadetblue", "darkgoldenrod", "black", "darkolivegreen", "firebrick4", "darkorchid4", "darkslategray", "mistyrose")
plot(allcells_x, allcells_y, col = mycolors[ as.numeric(mymodules[[1]]$module) ], pch=15, cex=2)
As an example I have done an analysis with all Australian (and some New Guinean) lycopods, the dataset I mentioned in the previous post. It plots as follows.
There are, of course, a few issues here. The analysis produced six modules, but three of them, the green, orange and light blue ones, consist of only two, one and one cells, respectively, and they seem biologically unrealistic. They may be artifacts of not having cleaned the data as well as I would for an actual study, or represent some kind of edge effect. The remaining three modules are clearly more meaningful. Although they contain some outlier cells, we can start to interpret them as potentially representing tropical (red), temperate (yellow), and subalpine/alpine (dark blue) assemblies of species, respectively.
Despite the less than perfect results I hope the example shows how easy it is to do such a Modularity Analysis, and if due diligence is done to the spatial data, as we would do in an actual study, I would also expect the results to become cleaner.
Saturday, March 17, 2018
Bioregionalisation part 5: Cleaning point distribution data in R
I should finally complete my series on bioregionalisation. What is missing is a post on how to do a network (Modularity) analysis in R. But first I thought I would write a bit about how to efficiently do some cleaning of point distribution data in R. As often I write this because it may be useful to somebody who finds it via search engine, but also because I can then look it up myself if I need it after not having done it for months.
The assumption is that we start our spatial or biogeographic analyses by obtaining point distribution data by querying e.g. for the genus or family that we want to study on an online biodiversity database or aggregator such as GBIF or Atlas of Living Australia. We download the record list in CSV format and now presumably have a large file with many columns, most of them irrelevant to our interests.
One problem that we may find is that there are numerous cases of records occurring in implausible locations. They may represent geospatial data entry errors such as land plants supposedly occurring in the ocean, or vouchers collected from plants in botanic gardens where the databasers fo some reason entered the garden's coordinates instead of those of the source location , or other outliers that we suspect to be misidentifications. What follows assumes that this at least has been done already (and it is hard to automate anyway), but we can use R to help us with a few other problems.
We start up R and begin by reading in our data, in this case all lycopod records downloaded from ALA. (One of the advantages about that group is that very few of them are cultivated in botanic gardens, and I did not want to do that kind of data clean-up for a blog post.)
Check again unique(mydata$species) to see if the situation has improved. If there are instances of name variants or outdated taxonomy that need to be corrected, that is surprisingly easy with a command along the following lines:
Although we assume that we had checked for geographic outliers, we may now still want to limit our analysis to a specific area. In my case I want to get rid of non-Australian records, so I remove every record outside of a box of 9.5 to 44.5 degrees south and 111 to 154 degrees east around the continent. Although it turns out that this left parts of New Guinea in that is fine with me for present purposes, we don't want to over-complicate this now.
The assumption is that we start our spatial or biogeographic analyses by obtaining point distribution data by querying e.g. for the genus or family that we want to study on an online biodiversity database or aggregator such as GBIF or Atlas of Living Australia. We download the record list in CSV format and now presumably have a large file with many columns, most of them irrelevant to our interests.
One problem that we may find is that there are numerous cases of records occurring in implausible locations. They may represent geospatial data entry errors such as land plants supposedly occurring in the ocean, or vouchers collected from plants in botanic gardens where the databasers fo some reason entered the garden's coordinates instead of those of the source location , or other outliers that we suspect to be misidentifications. What follows assumes that this at least has been done already (and it is hard to automate anyway), but we can use R to help us with a few other problems.
We start up R and begin by reading in our data, in this case all lycopod records downloaded from ALA. (One of the advantages about that group is that very few of them are cultivated in botanic gardens, and I did not want to do that kind of data clean-up for a blog post.)
rawdata <- read.csv("Lycopodiales.csv", sep=",", na.strings = "", header=TRUE, row.names=NULL)We now want to remove all records that lack any of the data we need for spatial and biogeographic analyses, i.e. identification to the species level, latitude and longitude. Other filtering may be desired, e.g. of records with too little geocode precision, but we will leave it at that for the moment. In my case the relevant columns are called genus, specificEpithet, decimalLatidue, and decimalLongitude, but that may of course be different in other data sources and require appropriate adjustment of the commands below.
rawdata <- rawdata[!(is.na(rawdata$decimalLatitude) | rawdata$decimalLatitude==""), ]All the records missing those data should be gone now. Next we make a new data frame containing only the data we are actually interested in.
rawdata <- rawdata[!(is.na(rawdata$decimalLongitude) | rawdata$decimalLongitude==""), ]
rawdata <- rawdata[!(is.na(rawdata$genus) | rawdata$genus==""), ]
rawdata <- rawdata[!(is.na(rawdata$specificEpithet.1) | rawdata$specificEpithet.1==""), ]
lat <- rawdata$decimalLatitudeUnfortunately at this stage there are still records that we may not want for our analysis, but they can mostly be recognised by having more than the two usual name elements of genus name and specific epithet: hybrids (something like "Huperzia prima x secunda" or "Huperzia x tertia") and undescribed phrase name taxa that may or may not actually be distinct species ("Lycopodiella spec. Mount Farewell"). At the same time we may want to check the list of species in our data table with unique(mydata$species) to see if we recognise any other problems that actually have two name elements, such as "Lycopodium spec." or "Lycopodium Undesignated". If there are any of those, we place them into a vector:
long <- rawdata$decimalLongitude
species <- paste( as.character(rawdata$genus), as.character(rawdata$specificEpithet.1, sep=" ") )
mydata <- data.frame(species, lat, long)
mydata$species <- as.character(mydata$species)
kickout <- c("Lycopodium spec.", "Lycopodium Undesignated")Then we loop through the data to get rid of all these problematic entries.
myflags <- rep(TRUE, length(mydata[,1]))If there is no 'kickout' vector for undesirable records with two name elements, we do the same but adjust the if command accordingly to not expect its existence.
for (i in 1:length(myflags))
{
if ( (length(strsplit(mydata$species[i], split=" ")[[1]]) != 2) || (mydata$species[i]) %in% kickout )
{
myflags[i] <- FALSE
}
}
mydata <- mydata[myflags, ]
Check again unique(mydata$species) to see if the situation has improved. If there are instances of name variants or outdated taxonomy that need to be corrected, that is surprisingly easy with a command along the following lines:
mydata$species[mydata$species == "Outdatica fastigiata"] = "Valida fastigiata"In that way we can efficiently harmonise the names so that one species does not get scored as two just because some specimens still have an outdated or misspelled name.
Although we assume that we had checked for geographic outliers, we may now still want to limit our analysis to a specific area. In my case I want to get rid of non-Australian records, so I remove every record outside of a box of 9.5 to 44.5 degrees south and 111 to 154 degrees east around the continent. Although it turns out that this left parts of New Guinea in that is fine with me for present purposes, we don't want to over-complicate this now.
mydata <- mydata[mydata$long<154, ]At this stage we may want to save the cleaned up data for future use, just in case.
mydata <- mydata[mydata$long>111, ]
mydata <- mydata[mydata$lat>(-44.5), ]
mydata <- mydata[mydata$lat<(-9.5), ]
write.table(mydata, file = "Lycopodiales_records_cleaned.csv", sep=",")And now, finally, we can actually turn the point distribution data into grid cells and conduct a network analysis, but that will be the next (and final) post of the series.
Saturday, January 27, 2018
Bioregionalisation part 3: clustering with Biodiverse
Biodiverse is a software for spatial analysis of biodiversity, in particular for calculating diversity scores for regions and for bioregionalisation. As mentioned in previous posts, the latter is done with clustering. Biodiverse is freely available and extremely powerful, just about the only minor issues are that the terminology used can sometimes be a bit confusing, and it is not always easy to intuit where to find a given function. As so often, a post like this might also help me to remember some detail when getting back to a program after a few months or so...
The following is about how to do bioregionalisation analysis in Biodiverse. First, the way I usually enter my spatial data is as one line per sample. So if you have coordinates, the relevant comma separated value file could look something like this:
Navigate to your file and select it. Note where you can choose the format of the data file in the lower right corner. Then click 'next'.
The following dialogue can generally be ignored, click 'next' once more.
But the third dialogue box is crucial. Here you need to tell Biodiverse how to interpret the data. The species (or other taxa) need to be interpreted as 'label', which is Biodiversian for the things that are found in regions. The coordinates need to be interpreted as 'group', the Biodiversian term for information that defines regions. For the grouping information the software also needs to be told if it is dealing with degrees for example, and what the size of the cells is supposed to be. In this case we have degrees and want one degree squared cells, but we could just as well have meters and want 100,000 m x 100,000 m cells.
After this we find ourselves confronted with yet another dialogue box and learn that despite telling Biodiverse which column is lat and which one is long it still doesn't understand that the stuff we just identified as long is meant to be on the x axis of a map. Arrange the two on the right so that long is above lat, and you are ready to click OK.
The result should be something like this: under a tab called 'outputs' we now have our input, i.e. our imported spatial data.
Double-clicking on the name of this dataset will produce another tab in which we can examine it. Clicking on a species name will mark its distribution on the map below. Clicking onto a cell on the map will show how similar other cells are to it in their species content. This will, of course, be much less clear if your cells are just region names, because in that case they will not be plotted in a nice two-dimensional map.
Now it is time to start our clustering analysis. Select 'Analyses -> cluster' from the menu. A third tab will open where you can select analysis parameters. Here I have chosen S2 dissimilarity as the metric. If there are ties during clustering it makes sense to break them by maximising endemism (because that is the whole point of the analysis anyway), so I set it to use Corrected Weighted Endemism first and then Weighted Endemism next if the former still does not resolve the situation. One could use random tie-breaks, but that would mean an analysis is not reproducible. All other settings were left as defaults.
After the analysis is completed, you can have the results displayed immediately. Alternatively, you can always go back to the first tab, where you will now find the analysis listed, and double-click it to get the display.
As we can see there is a dendrogram on the right and a map on the left. There are two ways of exploring nested clusters: Either change the number of clusters in the box at the bottom, or drag the thick blue line into a different position on the dendrogram; I find the former preferable. Note that if you increase the number too much Biodiverse will at a certain point run out of colours to display the clusters.
The results map is good, but we you may want to use the cluster assignments of the cells for downstream analyses in different software or simply to produce a better map somewhere else. How do you export the results? Not from the display interface. Instead, go back to the outputs tab, click the relevant analysis name, and then click 'export' on the right.
You now have an interface where you can name your output file, navigate to the desired folder, and select the number of clusters to be recognised under the 'number of groups' parameter on the left.
The reward should be a csv file like the following, where 'ELEMENT' is the name of each cell and 'NAME' is the column indicating what cluster each cell belongs to.
Again, very powerful, only have to keep in mind that your bioregions, for example, are variously called clusters, groups, and NAME depending on what part of the program you are dealing with.
The following is about how to do bioregionalisation analysis in Biodiverse. First, the way I usually enter my spatial data is as one line per sample. So if you have coordinates, the relevant comma separated value file could look something like this:
species,lat,longTo use equal area grid cells you may have reprojected the data so that lat and long values are in meters, but the format is of course the same. Alternatively, you may have only one column for the spatial information if your cells are not going to be coordinate-based but, for example, political units or bioregions:
Planta vulgaris,-26.45,145.29
Planta vulgaris,-27.08,144.88
...
species,stateJust for the sake of completeness, different formats such as a tsv would also work. Now to the program itself. You are running Biodiverse and choose 'Basedata -> Import' from the menus.
Planta vulgaris,Western Australia
Planta vulgaris,Northern Territory
...
Navigate to your file and select it. Note where you can choose the format of the data file in the lower right corner. Then click 'next'.
The following dialogue can generally be ignored, click 'next' once more.
But the third dialogue box is crucial. Here you need to tell Biodiverse how to interpret the data. The species (or other taxa) need to be interpreted as 'label', which is Biodiversian for the things that are found in regions. The coordinates need to be interpreted as 'group', the Biodiversian term for information that defines regions. For the grouping information the software also needs to be told if it is dealing with degrees for example, and what the size of the cells is supposed to be. In this case we have degrees and want one degree squared cells, but we could just as well have meters and want 100,000 m x 100,000 m cells.
After this we find ourselves confronted with yet another dialogue box and learn that despite telling Biodiverse which column is lat and which one is long it still doesn't understand that the stuff we just identified as long is meant to be on the x axis of a map. Arrange the two on the right so that long is above lat, and you are ready to click OK.
The result should be something like this: under a tab called 'outputs' we now have our input, i.e. our imported spatial data.
Double-clicking on the name of this dataset will produce another tab in which we can examine it. Clicking on a species name will mark its distribution on the map below. Clicking onto a cell on the map will show how similar other cells are to it in their species content. This will, of course, be much less clear if your cells are just region names, because in that case they will not be plotted in a nice two-dimensional map.
Now it is time to start our clustering analysis. Select 'Analyses -> cluster' from the menu. A third tab will open where you can select analysis parameters. Here I have chosen S2 dissimilarity as the metric. If there are ties during clustering it makes sense to break them by maximising endemism (because that is the whole point of the analysis anyway), so I set it to use Corrected Weighted Endemism first and then Weighted Endemism next if the former still does not resolve the situation. One could use random tie-breaks, but that would mean an analysis is not reproducible. All other settings were left as defaults.
After the analysis is completed, you can have the results displayed immediately. Alternatively, you can always go back to the first tab, where you will now find the analysis listed, and double-click it to get the display.
As we can see there is a dendrogram on the right and a map on the left. There are two ways of exploring nested clusters: Either change the number of clusters in the box at the bottom, or drag the thick blue line into a different position on the dendrogram; I find the former preferable. Note that if you increase the number too much Biodiverse will at a certain point run out of colours to display the clusters.
The results map is good, but we you may want to use the cluster assignments of the cells for downstream analyses in different software or simply to produce a better map somewhere else. How do you export the results? Not from the display interface. Instead, go back to the outputs tab, click the relevant analysis name, and then click 'export' on the right.
You now have an interface where you can name your output file, navigate to the desired folder, and select the number of clusters to be recognised under the 'number of groups' parameter on the left.
The reward should be a csv file like the following, where 'ELEMENT' is the name of each cell and 'NAME' is the column indicating what cluster each cell belongs to.
Again, very powerful, only have to keep in mind that your bioregions, for example, are variously called clusters, groups, and NAME depending on what part of the program you are dealing with.
Wednesday, July 19, 2017
How to use the reference manager Zotero
(Updated 19 July 2017 regarding Zotero 5.)
Inspired by an eMail exchange with a colleague I thought I would write a longer post on how to use the open source reference manager Zotero. Obviously all the information will in some form be available in its documentation, but at least to me the things I really need to look up are often the needle in the haystack of the obvious and the irrelevant. So here is what I think is what one needs to know to start using Zotero in what is hopefully a logical order.
All of this is based on Zotero 4, as I have not yet used the newest, version 5. From version 5 the standalone program is required instead of Zotero running through the browser alone. See the relevant comment under installation below.
How it works
Zotero is available for Win, Mac and Linux and for LibreOffice or MS Word. It integrates into the browser (I use Firefox, but it also seems to work with Chrome, Safari and Opera), and the usual way to use it is through the browser. This means that the browser has to be open at the same time as the word processor, but there is also a standalone version that I am not familiar with.
Installation
To be honest, this is the only point on which I am a bit confused at the moment because it has been a bit since I installed one of my instances. There are three items that may have to be installed: Zotero itself on the download page, for which there are specific instructions. Then there is the "connector" for the browser you have. Finally, it may be necessary to install a word processor plug-in for your browser. What confuses me is that I seem to remember only installing the latter two last time, so either I misremember or something has changed with the newest Zotero version (?).
Either way, while the installation of Zotero into the browser itself is easy, I have noticed that sometimes the word processor plug-in does not take on the first attempt. In that case I merely repeated the installation and restarted everything, and then it worked.
Update: The colleague who has now started using Zotero has, of course, installed the newest version and kindly adds the following:
Using Zotero in the browser and building your reference library
When you have Zotero installed there are two new buttons in the browser: a "Z" that opens your reference library and a symbol right next to it that you can click to import a journal article into that library. Given that the library will at first be empty let's look at that latter function first.
For example, I may have found a journal article through a Google Scholar search. Ideally, I am now looking at the abstract or HTML fulltext on the journal website, because that page will have all the metadata I want. I now click on the paper symbol to the right of the Z, and Zotero automatically grabs all the fields it can find and saves a new entry into my reference library; if it can get a PDF it will even download that.
Now I click on the Z button to bring up my library, if I didn't have it open yet. In some cases I may notice that something went wrong. The typical scenarios are that the title of the paper is in Title Case or ALL CAPS. This is easily rectified by right-clicking on the title field and selecting "sentence case".
If something needs to be edited manually we can do so by left-clicking onto the relevant field. For paper titles, the usual problems would be having to re-capitalise names after correcting for Title Case or adding the HTML tags for italics round organism names. In the present case, however, I find that the title contains HTML codes for single quotation marks instead of the actual quotation mark characters, so I quickly correct that. Manual entry is, of course, also possible for an entire reference, for example if it isn't available online. In that case simply click on the plus in the green circle and select the appropriate publication type.
So much for importing references. It is also possible to bulk-import from the Google Scholar search results, but I would not recommend that as Google sometimes mixes up the metadata.
The style repository
The first time we try to add a reference to a manuscript, we are asked what reference style should be used. Zotero comes with only a few standard styles installed, but many more are available at the Zotero style repository. One of the in my eyes few downsides of Zotero is that it has less styles than Endnote, but often it is possible to get the relevant one under a different name. If, for example, you are preparing a manuscript for PhytoTaxa the ZooTaxa style should serve just as well.
Installing a new style is as easy as finding it in the style repository, clicking on its name, and confirming that it should be installed.
Using Zotero in the word processor
Again, note that the browser needs to be running while we are adding references to a paper. The following assumes LibreOffice, but except for where to find the buttons everything is the same in MS Word.
In LibreOffice you will have new buttons for inserting and editing references, for inserting the reference list, and for changing document settings, in particular the reference style. To insert a reference, click on the button that seems to read r."Z. You can now enter an author name or even just a word from the title, as in my example here, and Zotero will suggest anything that fits.
Another downside of Zotero, at least as of version 4 which I am still using, is that it doesn't do a reference like "Bronzati (2017)". Instead you can either have "(Bronzati 2017)" or reduce the reference to "(2017)". For this click on the reference in the field where you were asked to select it (if you have already entered it simply use the edit reference button showing r." and pencil) and select "suppress author". Then you have to type the author name(s) yourself outside of the brackets, which is obviously a bit annoying.
Once we have added a few references, we obviously need to add the reference list. This is as easy as clicking the third button in the Zotero field. The only others that are usually important are the two arrows (refresh) to update the reference list (although it does so automatically when the document is reloaded) and the cogwheel (document preferences) that allows changing the reference style across the document.
In LibreOffice I have sometimes found that adding or updating references changes the format of the entire paragraph they are embedded in. This seems to happen if the default text style is at variance with the text format actually used in the manuscript. Selecting a piece of manuscript text and setting the default style to fit its format has always rectified the situation for me.
Syncing
It is useful to get an account at the Zotero website and use it to sync one's reference library across computers. Again, this works cross-platform. I do it between a Windows computer at work and my personal Linux computer at home. Note, however, that it only syncs the metadata, not any fulltext PDFs that have been saved.
To sync, go into the browser and click on the Z symbol to open Zotero. Now click the cogwheel and select preferences. The preferences window has a sync tab where you can enter your username and password. Do the same on two computers and they should share their reference libraries.
Inspired by an eMail exchange with a colleague I thought I would write a longer post on how to use the open source reference manager Zotero. Obviously all the information will in some form be available in its documentation, but at least to me the things I really need to look up are often the needle in the haystack of the obvious and the irrelevant. So here is what I think is what one needs to know to start using Zotero in what is hopefully a logical order.
All of this is based on Zotero 4, as I have not yet used the newest, version 5. From version 5 the standalone program is required instead of Zotero running through the browser alone. See the relevant comment under installation below.
How it works
Zotero is available for Win, Mac and Linux and for LibreOffice or MS Word. It integrates into the browser (I use Firefox, but it also seems to work with Chrome, Safari and Opera), and the usual way to use it is through the browser. This means that the browser has to be open at the same time as the word processor, but there is also a standalone version that I am not familiar with.
Installation
To be honest, this is the only point on which I am a bit confused at the moment because it has been a bit since I installed one of my instances. There are three items that may have to be installed: Zotero itself on the download page, for which there are specific instructions. Then there is the "connector" for the browser you have. Finally, it may be necessary to install a word processor plug-in for your browser. What confuses me is that I seem to remember only installing the latter two last time, so either I misremember or something has changed with the newest Zotero version (?).
Either way, while the installation of Zotero into the browser itself is easy, I have noticed that sometimes the word processor plug-in does not take on the first attempt. In that case I merely repeated the installation and restarted everything, and then it worked.
Update: The colleague who has now started using Zotero has, of course, installed the newest version and kindly adds the following:
The new version of Zotero needs the standalone program installed. This is because Mozilla has dropped the engine that supported a lot of extensions (like Zotero). It was deemed that allowing the browser to carry out low-level functions on the host computer introduced inherent vulnerabilities, and so Firefox versions after 48 have very much restricted what the browser is allowed to do (no longer can it communicate directly with databases, and carry out a lot of file handling functions). The problem is not restricted to Zotero, for example Gnome desktop extensions used this functionality and have also had to change the way they do things. The long and the short is that nowadays you need the standalone application installed.
Using Zotero in the browser and building your reference library
When you have Zotero installed there are two new buttons in the browser: a "Z" that opens your reference library and a symbol right next to it that you can click to import a journal article into that library. Given that the library will at first be empty let's look at that latter function first.
 |
| The Zotero buttons in the browser |
 |
| Viewing paper abstract |
 |
| A reference has been added to the library |
So much for importing references. It is also possible to bulk-import from the Google Scholar search results, but I would not recommend that as Google sometimes mixes up the metadata.
The style repository
The first time we try to add a reference to a manuscript, we are asked what reference style should be used. Zotero comes with only a few standard styles installed, but many more are available at the Zotero style repository. One of the in my eyes few downsides of Zotero is that it has less styles than Endnote, but often it is possible to get the relevant one under a different name. If, for example, you are preparing a manuscript for PhytoTaxa the ZooTaxa style should serve just as well.
Installing a new style is as easy as finding it in the style repository, clicking on its name, and confirming that it should be installed.
 |
| Selecting a reference style |
Again, note that the browser needs to be running while we are adding references to a paper. The following assumes LibreOffice, but except for where to find the buttons everything is the same in MS Word.
In LibreOffice you will have new buttons for inserting and editing references, for inserting the reference list, and for changing document settings, in particular the reference style. To insert a reference, click on the button that seems to read r."Z. You can now enter an author name or even just a word from the title, as in my example here, and Zotero will suggest anything that fits.
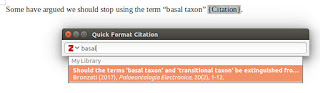 |
| Adding a reference to a manuscript |
 |
| Author names outside of brackets have to be added manually |
In LibreOffice I have sometimes found that adding or updating references changes the format of the entire paragraph they are embedded in. This seems to happen if the default text style is at variance with the text format actually used in the manuscript. Selecting a piece of manuscript text and setting the default style to fit its format has always rectified the situation for me.
Syncing
It is useful to get an account at the Zotero website and use it to sync one's reference library across computers. Again, this works cross-platform. I do it between a Windows computer at work and my personal Linux computer at home. Note, however, that it only syncs the metadata, not any fulltext PDFs that have been saved.
To sync, go into the browser and click on the Z symbol to open Zotero. Now click the cogwheel and select preferences. The preferences window has a sync tab where you can enter your username and password. Do the same on two computers and they should share their reference libraries.
Sunday, June 11, 2017
How the sausage is made: peer reviewing edition
One of the aspects of working as a scientist that I find most intriguing is peer reviewing each other's work. The main issue is that while how to write the actual manuscripts is explicitly and formally taught and further supported by style guides, helpful books and journals' instructions to authors, there is much less formal instruction on how to write a reviewer's report.
Essentially one is limited to (1) relatively vague journals' instructions to reviewers usually on the lines of "be constructive" or "be charitable", (2) deducing what matters to the editor from the questions asked in the reviewer's report form, and (3) emulating the style of the comments one receives on one's own papers. Apart from generic, often system-generated thank you messages there is generally no feedback on whether and to what degree the editors found my reviewer's reports appropriate and helpful or on how they compared with other reports.
In other words, most of it is learning by doing; after years of practice I now have a good overview of what reviewer reports in my field look like, but as a beginner I had very little to go by.
It is then no surprise that the style and tone in which reviewers in my field write their reports can differ quite a lot from person to person. There is a general pattern of first discussing general issues and broad suggestions and then minor suggestions line-by-line on phrasing, word choice or typos, and there is clearly the expectation of being reasonable and civil. But:
First, I like to print the manuscript - I am old-fashioned like that. I try to begin reading it fairly soon after I accept the job, and for obvious reasons I also try to read through more or less over one day. Often I will read when I need a break from some other task like computer work, on a bus or in the evening at home.
Already on the first read I attempt to thoroughly mark everything I notice. Using a red or blue pen I mark minor issues a bit like a teacher correcting a dictation, while making little notes on the margins where I have more general concerns ("poorly explained", "circular", "what about geographic outliers?").
Usually the following day I order my thoughts on the manuscript and start a very rough report draft by first typing out all the minor suggestions. (I would prefer to use tracked changes on a Word document for that, but unfortunately we generally only get a PDF, and I find annotating those even more tedious than just writing things out.) Then I start on the general concerns, if any, merely by writing single sentences on each point but do not expand just then.
In particular if the study is valuable but has some weaknesses I prefer to sleep over it at this stage for 2-3 nights or, if the task has turned out to be a bit unpleasant, even a few days more, and then look at it again with fresh eyes. That helps me to avoid being overly negative; in fact I tend to start out rather bluntly and then, with some distance, rephrase and expand my comments to be more polite and constructive.
That being said, if the manuscript is nearly flawless or totally unsalvageable I usually finish my review very quickly. If I remember correctly my record is something like 45 min after being invited to review, because the study was just that deeply flawed. In that case I saw no reason to spend a lot of time on trying to develop a list of minor suggestions.
More generally I have over the years come to the conclusion that it cannot be the role of a peer reviewer to check if all papers in the reference list have really been cited or to suggest language corrections in each paragraph, although some colleagues seem to get a kick out of that. If there are more than a handful of language issues I would simply say that the language needs work instead of pointing out each instance, and if there are issues with the references I would suggest the authors consider using a reference manager such as Zotero, done. Really from my perspective the point of peer review is to check if the science is sound, and everything else is at best a distant secondary concern.
At any rate, after having slept over the manuscript a bit I will return to it and write the general comments out into more fluent text. I aim to do the usual sandwich: start with a positive paragraph that summarises the main contribution made by the manuscript and what I particularly like about it. If necessary, this is followed by something to the effect of "nonetheless I have some concerns" or "unfortunately, some changes are required before this useful contribution can be published".
Then comes the major stuff that I would suggest to change, delete or add, including in each case with a concrete recommendation of what could be done to improve the manuscript. I follow a logical order through the text but usually end with what I consider most important, or repeat that point if it was already covered earlier. To end the general comments on something positive I will have another paragraph stressing how valuable the manuscript would be, that I hope it will ultimately be published, etc. Even if I feel I have to suggest rejection I try to stress a positive element of the work.
Finally, and as mentioned above, there is the list of minor suggestions. Most other reviewers I have run into seem to structure their reports similarly.
When submitting the report, however, one does not only have to provide the text I have discussed so far, although it is certainly the most useful from the authors' perspective. No, nearly all journals have a field of "reviewer blind comments to the editor", which I rarely find necessary to use, and a number of questions that the reviewer has to answer. The latter are typically on the following lines:
Essentially one is limited to (1) relatively vague journals' instructions to reviewers usually on the lines of "be constructive" or "be charitable", (2) deducing what matters to the editor from the questions asked in the reviewer's report form, and (3) emulating the style of the comments one receives on one's own papers. Apart from generic, often system-generated thank you messages there is generally no feedback on whether and to what degree the editors found my reviewer's reports appropriate and helpful or on how they compared with other reports.
In other words, most of it is learning by doing; after years of practice I now have a good overview of what reviewer reports in my field look like, but as a beginner I had very little to go by.
It is then no surprise that the style and tone in which reviewers in my field write their reports can differ quite a lot from person to person. There is a general pattern of first discussing general issues and broad suggestions and then minor suggestions line-by-line on phrasing, word choice or typos, and there is clearly the expectation of being reasonable and civil. But:
- Some reviewers may summarise the manuscript abstract-style before they start their evaluation, while others assume that the editor does not need that information given that they have the actual abstract of the paper available to them.
- Some stick to evaluating the scientific accuracy of the paper, while others obsess about wording and phrasing and regularly ask authors who are native speakers of English to have a native speaker of English check their manuscript.
- Some stick to judging whether the analysis chosen by the authors is suitable to answer the study question, while others see an opportunity to suggest the addition of five totally irrelevant analyses just because they happen to know they are possible. And sometimes they recommend cutting another 2,000 words from the text despite suggesting those additions, as if those would come without text.
- Some unashamedly use the reviewer's report for self-promotion by suggesting that some of their own publications be cited, relevant or not.
- Some use a professional tone and make constructive suggestions on the particular manuscript in question, but others apparently cannot help disparaging the authors themselves. Luckily that behaviour is rare.
- Some write one paragraph even when they recommend major revision (meaning they could have been more explicit about what and how to revise), others write six pages of suggestions even when their recommendation is rather positive.
First, I like to print the manuscript - I am old-fashioned like that. I try to begin reading it fairly soon after I accept the job, and for obvious reasons I also try to read through more or less over one day. Often I will read when I need a break from some other task like computer work, on a bus or in the evening at home.
Already on the first read I attempt to thoroughly mark everything I notice. Using a red or blue pen I mark minor issues a bit like a teacher correcting a dictation, while making little notes on the margins where I have more general concerns ("poorly explained", "circular", "what about geographic outliers?").
Usually the following day I order my thoughts on the manuscript and start a very rough report draft by first typing out all the minor suggestions. (I would prefer to use tracked changes on a Word document for that, but unfortunately we generally only get a PDF, and I find annotating those even more tedious than just writing things out.) Then I start on the general concerns, if any, merely by writing single sentences on each point but do not expand just then.
In particular if the study is valuable but has some weaknesses I prefer to sleep over it at this stage for 2-3 nights or, if the task has turned out to be a bit unpleasant, even a few days more, and then look at it again with fresh eyes. That helps me to avoid being overly negative; in fact I tend to start out rather bluntly and then, with some distance, rephrase and expand my comments to be more polite and constructive.
That being said, if the manuscript is nearly flawless or totally unsalvageable I usually finish my review very quickly. If I remember correctly my record is something like 45 min after being invited to review, because the study was just that deeply flawed. In that case I saw no reason to spend a lot of time on trying to develop a list of minor suggestions.
More generally I have over the years come to the conclusion that it cannot be the role of a peer reviewer to check if all papers in the reference list have really been cited or to suggest language corrections in each paragraph, although some colleagues seem to get a kick out of that. If there are more than a handful of language issues I would simply say that the language needs work instead of pointing out each instance, and if there are issues with the references I would suggest the authors consider using a reference manager such as Zotero, done. Really from my perspective the point of peer review is to check if the science is sound, and everything else is at best a distant secondary concern.
At any rate, after having slept over the manuscript a bit I will return to it and write the general comments out into more fluent text. I aim to do the usual sandwich: start with a positive paragraph that summarises the main contribution made by the manuscript and what I particularly like about it. If necessary, this is followed by something to the effect of "nonetheless I have some concerns" or "unfortunately, some changes are required before this useful contribution can be published".
Then comes the major stuff that I would suggest to change, delete or add, including in each case with a concrete recommendation of what could be done to improve the manuscript. I follow a logical order through the text but usually end with what I consider most important, or repeat that point if it was already covered earlier. To end the general comments on something positive I will have another paragraph stressing how valuable the manuscript would be, that I hope it will ultimately be published, etc. Even if I feel I have to suggest rejection I try to stress a positive element of the work.
Finally, and as mentioned above, there is the list of minor suggestions. Most other reviewers I have run into seem to structure their reports similarly.
When submitting the report, however, one does not only have to provide the text I have discussed so far, although it is certainly the most useful from the authors' perspective. No, nearly all journals have a field of "reviewer blind comments to the editor", which I rarely find necessary to use, and a number of questions that the reviewer has to answer. The latter are typically on the following lines:
- Is the language acceptable or is revision required?
- Are the conclusions sound and do they follow logically from the results?
- Are all the tables and figures necessary?
Thursday, March 16, 2017
Some very, very basic notes on paper writing
The following are just a few notes on manuscript or student report writing for my area of science, which I would circumscribe as plant systematics, biogeography and evolutionary biology. I will probably at some point use this or a revised version for other purposes, but thought it might be interesting to blog about.
As should be well known, the typical research paper (and a student report mirrored after it) in my area has, in this order, a title, author names and contact info, an abstract, key words, introduction, materials and methods, results, discussion, optionally conclusions (or they may be part of the discussion), acknowledgements, reference list, figure legends, tables, and potentially appendices or supplementary data. I will only deal with some of these for now. The main text, i.e. without reference list, should probably not be longer than c. 5,000 words, and the shorter the better.
Abstract
The abstract should summarise all the other sections in really abbreviated form: what is the paper about, what main methods were used, what are the main results, and what do they mean. Do not cite references in the abstract, and do not provide taxonomic authorities after plant names. They are provided on the first mention (and only on the first mention) of a name in the main text.
Introduction
This section provides background information, describes the question or problem, and ends with aims of the study. Ideally start with general, well-established, and unproblematic claims ("biodiversity loss is accelerating", "genomic data have become increasingly available for use in phylogenetic studies", etc.) and move relatively quickly to the problem ("guidance is needed to prioritise conservation planning", "but analysis of the large amounts of data produced by high-throughput sequencing is computationally challenging"). Do not begin with your study group, unless the paper is a purely taxonomic or phylogenetic one, as this will not draw in as many readers; the introduction of the study group should come after the general question has been established.
Obviously the introduction needs to be full of references, and all but the most widely accepted statements should be followed by at least one of them.
The aims should follow logically from the questions or problems and need to tie in logically with the methods, results and discussion. They can be formulated as hypotheses or questions, but should not be too vague. Think "we will test if patterns agree with those postulated by Smith (1980)" instead of "we want to explore the patterns".
Methods
Be as concise as you can. Cite software, tests and methods, but not the most basic ones; PCR or t-tests, for example, can be considered sufficiently established. Start with the materials (sampling strategy etc.), then work logically through the lab and analysis pipeline.
Results
This section presents the results and nothing more. Any sentence that interprets the results or explains them does not belong here but into the discussion. References do not belong here. Explanation of how something was done does not belong here but into the methods.
On the other hand, all observations need to be stated explicitly in the main text of the results section, with a reference to the figure or table where they are presented. The figure legend itself should, in turn, be very concise and merely provide enough information to understand the figure, but it should not restate what the reader can see in the figure just above said legend anyway. For example, the figure legend might say "A, map of Australia showing endemism hotspots", but something like "hotspots are found in the southwest and southeast" is superfluous here and goes into the main text: "endemism hotspots are found in the southwest and southeast (Fig. 2A)".
Discussion
Perhaps the hardest part to write, it explains what the results mean and how they fit into the wider context. It may also end on suggestions for further study. The main problem for beginners is to avoid repeating what was already said in the introduction and results sections.
The discussion should again have lots of references - not because we always need to cite lots of papers per se, but because any paragraph in the discussion that does not have at least one reference can be considered under suspicion of either belonging into the results section or being mere padding.
Acknowledgements
Collaborators, funding sources, peer reviewers. For student reports, one would expect the supervisor(s) to be mentioned. It is not clear to me why many people feel the need to initialise the names of those who they are thanking, as writing them out makes it easier to identify them, especially if they have common family names.
Figures
I haven't really made a tally, but the typical article would probably have between two and six figures, after that it becomes excessive.
Journals in my area expect the manuscript text and figures to be uploaded as separate files, and they expect high quality file formats such as EPS for vector graphics and TIFs with lossless compression for bitmaps.
For exchanging a manuscript draft between co-authors, or a student report draft between student and supervisor, it is, however, probably best to insert figures into the manuscript file, because then your collaborator has less files to handle and to print. I would suggest the following approach:
Have a separate section for the figure legends after the references. This is as it should be for journal submission.
Produce EPS (vector) or JPGs (bitmap) of the figures to save on file size and insert each of them into the text directly above the relevant figure legend. Although Word allows for them, I would very strongly advise against using convoluted text boxes-inside-object boxes-inside-object boxes. A student report I once had to reformat froze my computer for several minutes when I tried to resize one of its "figures". Ultimately I had to export the page as a PDF and then export the PDF from Inkscape into yet another format, whereupon I reinserted the figure into the Word document. A journal would, of course, not accept anything like that anyway.
Select the wrapping option "in line with text". This will treat the figure like a character, meaning that it will remain anchored to its position relative to the text no matter what you do upwards of its position. If you use other wrapping options such as "on top of the text" the figures will float around freely, and changing line spacing or font size, or deleting paragraphs, will mess everything up to no end. I once dealt with a student report where three figures ended up on top of each other!
Do not use text boxes for the figure legends, or for tables, for that matter. Really, why would you? They can be normal text, just like the rest of the manuscript. In fact I have yet to see a use case for Word's text boxes in my line of work (PowerPoint is a different matter).
Language
There is no reason to use words like "whilst" where a simple "while" will do.
Often sentences can be simplified greatly; some people seem to have a penchant for writing something on the lines of "for pollination, it has been demonstrated that hummingbirds are more efficient", but the same could be said as "hummingbirds are more efficient pollinators" - saved us six words! Similarly, "in the literature it is documented that the sky is blue (Smith, 1980)" can be reduced to "the sky is blue (Smith, 1980)". And yes, I have seen sentences like these, particularly as a reviewer.
Small stuff
Do not use double spacers at the end of a sentence.
Do not have spacers at the end of a paragraph.
There needs to be a spacer between any measurement and its unit (5 km, 5 h), with the exception of temperatures (5ºC) and angles (5º).
This might seem a bit OCD, but believe me, you do not have to make some poor copy editor's life harder than it is.
As should be well known, the typical research paper (and a student report mirrored after it) in my area has, in this order, a title, author names and contact info, an abstract, key words, introduction, materials and methods, results, discussion, optionally conclusions (or they may be part of the discussion), acknowledgements, reference list, figure legends, tables, and potentially appendices or supplementary data. I will only deal with some of these for now. The main text, i.e. without reference list, should probably not be longer than c. 5,000 words, and the shorter the better.
Abstract
The abstract should summarise all the other sections in really abbreviated form: what is the paper about, what main methods were used, what are the main results, and what do they mean. Do not cite references in the abstract, and do not provide taxonomic authorities after plant names. They are provided on the first mention (and only on the first mention) of a name in the main text.
Introduction
This section provides background information, describes the question or problem, and ends with aims of the study. Ideally start with general, well-established, and unproblematic claims ("biodiversity loss is accelerating", "genomic data have become increasingly available for use in phylogenetic studies", etc.) and move relatively quickly to the problem ("guidance is needed to prioritise conservation planning", "but analysis of the large amounts of data produced by high-throughput sequencing is computationally challenging"). Do not begin with your study group, unless the paper is a purely taxonomic or phylogenetic one, as this will not draw in as many readers; the introduction of the study group should come after the general question has been established.
Obviously the introduction needs to be full of references, and all but the most widely accepted statements should be followed by at least one of them.
The aims should follow logically from the questions or problems and need to tie in logically with the methods, results and discussion. They can be formulated as hypotheses or questions, but should not be too vague. Think "we will test if patterns agree with those postulated by Smith (1980)" instead of "we want to explore the patterns".
Methods
Be as concise as you can. Cite software, tests and methods, but not the most basic ones; PCR or t-tests, for example, can be considered sufficiently established. Start with the materials (sampling strategy etc.), then work logically through the lab and analysis pipeline.
Results
This section presents the results and nothing more. Any sentence that interprets the results or explains them does not belong here but into the discussion. References do not belong here. Explanation of how something was done does not belong here but into the methods.
On the other hand, all observations need to be stated explicitly in the main text of the results section, with a reference to the figure or table where they are presented. The figure legend itself should, in turn, be very concise and merely provide enough information to understand the figure, but it should not restate what the reader can see in the figure just above said legend anyway. For example, the figure legend might say "A, map of Australia showing endemism hotspots", but something like "hotspots are found in the southwest and southeast" is superfluous here and goes into the main text: "endemism hotspots are found in the southwest and southeast (Fig. 2A)".
Discussion
Perhaps the hardest part to write, it explains what the results mean and how they fit into the wider context. It may also end on suggestions for further study. The main problem for beginners is to avoid repeating what was already said in the introduction and results sections.
The discussion should again have lots of references - not because we always need to cite lots of papers per se, but because any paragraph in the discussion that does not have at least one reference can be considered under suspicion of either belonging into the results section or being mere padding.
Acknowledgements
Collaborators, funding sources, peer reviewers. For student reports, one would expect the supervisor(s) to be mentioned. It is not clear to me why many people feel the need to initialise the names of those who they are thanking, as writing them out makes it easier to identify them, especially if they have common family names.
Figures
I haven't really made a tally, but the typical article would probably have between two and six figures, after that it becomes excessive.
Journals in my area expect the manuscript text and figures to be uploaded as separate files, and they expect high quality file formats such as EPS for vector graphics and TIFs with lossless compression for bitmaps.
For exchanging a manuscript draft between co-authors, or a student report draft between student and supervisor, it is, however, probably best to insert figures into the manuscript file, because then your collaborator has less files to handle and to print. I would suggest the following approach:
Have a separate section for the figure legends after the references. This is as it should be for journal submission.
Produce EPS (vector) or JPGs (bitmap) of the figures to save on file size and insert each of them into the text directly above the relevant figure legend. Although Word allows for them, I would very strongly advise against using convoluted text boxes-inside-object boxes-inside-object boxes. A student report I once had to reformat froze my computer for several minutes when I tried to resize one of its "figures". Ultimately I had to export the page as a PDF and then export the PDF from Inkscape into yet another format, whereupon I reinserted the figure into the Word document. A journal would, of course, not accept anything like that anyway.
Select the wrapping option "in line with text". This will treat the figure like a character, meaning that it will remain anchored to its position relative to the text no matter what you do upwards of its position. If you use other wrapping options such as "on top of the text" the figures will float around freely, and changing line spacing or font size, or deleting paragraphs, will mess everything up to no end. I once dealt with a student report where three figures ended up on top of each other!
Do not use text boxes for the figure legends, or for tables, for that matter. Really, why would you? They can be normal text, just like the rest of the manuscript. In fact I have yet to see a use case for Word's text boxes in my line of work (PowerPoint is a different matter).
Language
There is no reason to use words like "whilst" where a simple "while" will do.
Often sentences can be simplified greatly; some people seem to have a penchant for writing something on the lines of "for pollination, it has been demonstrated that hummingbirds are more efficient", but the same could be said as "hummingbirds are more efficient pollinators" - saved us six words! Similarly, "in the literature it is documented that the sky is blue (Smith, 1980)" can be reduced to "the sky is blue (Smith, 1980)". And yes, I have seen sentences like these, particularly as a reviewer.
Small stuff
Do not use double spacers at the end of a sentence.
Do not have spacers at the end of a paragraph.
There needs to be a spacer between any measurement and its unit (5 km, 5 h), with the exception of temperatures (5ºC) and angles (5º).
This might seem a bit OCD, but believe me, you do not have to make some poor copy editor's life harder than it is.
Thursday, December 8, 2016
How to set multiple calibration points in R's chronos function
As mentioned in this venue before, there are two fundamental ways of producing a time-calibrated phylogeny: either infer a Bayesian or Likelihood tree directly under a clock model, or infer a phylogram first and then force it into ultrametric shape afterwards. The second approach has the obvious advantage that you can use whatever tree you want, including e.g. parsimony trees. It also means that the topology is kept the same.
Some time ago I discussed how to use the program r8s, but as mentioned then it is only easily available on Mac and pretty much impossible to get to work on Windows. A more up-to-date alternative is the chronos function in the R package APE. And as mentioned in an even more recent post, it uses Penalised Likelihood with three different settings (relaxed, correlated or discrete) and an adjustable smoothing parameter lambda to time-calibrate the phylogeny.
The first few steps are well documented in APE's manual or on other online sources. Set your working directory, for example setwd("C:/Users/you/Documents/R stuff") . Load APE with library(ape). It needs to be installed, of course.
Next import your tree, either mytree <- read.tree("phylogram.tre") or mytree <- read.nexus("phylogram.tre") depending on whether it is saved in Newick or Nexus format.
Now if you want to merely specify the root age of the tree, for example as 15 million years, you have it easy. Just establish a calibration using APE's makeChronosCalibration function as described in the manual:
mycalibration <- makeChronosCalib(mytree, node="root", age.max=15)
You hand your tree, smoothing parameter, model choice and the calibration over to the chronos function:
mytimetree <- chronos(mytree, lambda = 1, model = "relaxed", calibration = mycalibration, control = chronos.control() )
Done! Now you can plot the tree or save it as usual. But what if you have several calibration points? The documentation does not provide an example of how to do that. It mentions an interactive mode in which the phylogram is displayed, you can click on one branch after the other, and each time you are asked to provide its minimum and maximum ages manually.
A very good discussion of the interactive mode was provided by a blogger called Justin Bagley a bit more than a year ago, but the so far first and only commenter raised the obvious issue:
So I spent some time figuring out how exactly we can build a data matrix of calibration points for chronos ourselves. Here is what you do.
Create a vector of nodes that you have calibrations for. I find it easiest to let APE find the node numbers itself by using the get Most Recent Common Ancestor function. For example, if you need to find the node where the ancestors of Osmunda and Doodia diverged, you would use getMRCA(mytree, tip = c("Osmunda", "Doodia")):
node <- c(
getMRCA(mytree, tip = c("Equisetum","Doodia") ),
getMRCA(mytree, tip = c("Osmunda","Doodia") ),
getMRCA(mytree, tip = c("Hymenophyllum","Doodia") )
)
So now we have defined three nodes. Next, minimum and maximum ages. The first is the root of my imaginary tree, so I want to set a maximum age as a wall for the whole tree depth but no minimum. The others are internal nodes calibrated with fossils, so they should be minimum ages but have no maximum. I tried to enter NA or NULL when there was no minimum or maximum but it didn't work, so the easiest thing to do is to specify 0 for no minimum and the root age for no maximum.
age.min <- c(
0,
280,
270
)
age.max <- c(
354,
354,
354
)
Finally, chronos expects a last column of the data matrix that is not currently used. Still it needs to have the right dimensions:
soft.bounds <- c(
FALSE,
FALSE,
FALSE
)
Now simply unite these four vectors into a single matrix:
mycalibration <- data.frame(node, age.min, age.max, soft.bounds)
The names of the vectors turn into the column names that chronos expects, and off we go: three calibration points. I hope some readers find this useful, and that it works as well for them as it did for me.
Some time ago I discussed how to use the program r8s, but as mentioned then it is only easily available on Mac and pretty much impossible to get to work on Windows. A more up-to-date alternative is the chronos function in the R package APE. And as mentioned in an even more recent post, it uses Penalised Likelihood with three different settings (relaxed, correlated or discrete) and an adjustable smoothing parameter lambda to time-calibrate the phylogeny.
The first few steps are well documented in APE's manual or on other online sources. Set your working directory, for example setwd("C:/Users/you/Documents/R stuff") . Load APE with library(ape). It needs to be installed, of course.
Next import your tree, either mytree <- read.tree("phylogram.tre") or mytree <- read.nexus("phylogram.tre") depending on whether it is saved in Newick or Nexus format.
Now if you want to merely specify the root age of the tree, for example as 15 million years, you have it easy. Just establish a calibration using APE's makeChronosCalibration function as described in the manual:
mycalibration <- makeChronosCalib(mytree, node="root", age.max=15)
You hand your tree, smoothing parameter, model choice and the calibration over to the chronos function:
mytimetree <- chronos(mytree, lambda = 1, model = "relaxed", calibration = mycalibration, control = chronos.control() )
Done! Now you can plot the tree or save it as usual. But what if you have several calibration points? The documentation does not provide an example of how to do that. It mentions an interactive mode in which the phylogram is displayed, you can click on one branch after the other, and each time you are asked to provide its minimum and maximum ages manually.
A very good discussion of the interactive mode was provided by a blogger called Justin Bagley a bit more than a year ago, but the so far first and only commenter raised the obvious issue:
I am trying to place several dozens of calibrations in a very large tree so the "clicking on the tree" step in the interactive mode is kind of tricky as I just cannot see the nodes especially the more derived ones.Also, it is just plain tedious for that many calibration points. What is more, I tried it myself earlier today and found that even when I provided a minimum age only and skipped on entering a maximum, the resulting calibration table would still have the maximum age identical to the minimum age - that is not how it is supposed to work.
So I spent some time figuring out how exactly we can build a data matrix of calibration points for chronos ourselves. Here is what you do.
Create a vector of nodes that you have calibrations for. I find it easiest to let APE find the node numbers itself by using the get Most Recent Common Ancestor function. For example, if you need to find the node where the ancestors of Osmunda and Doodia diverged, you would use getMRCA(mytree, tip = c("Osmunda", "Doodia")):
node <- c(
getMRCA(mytree, tip = c("Equisetum","Doodia") ),
getMRCA(mytree, tip = c("Osmunda","Doodia") ),
getMRCA(mytree, tip = c("Hymenophyllum","Doodia") )
)
So now we have defined three nodes. Next, minimum and maximum ages. The first is the root of my imaginary tree, so I want to set a maximum age as a wall for the whole tree depth but no minimum. The others are internal nodes calibrated with fossils, so they should be minimum ages but have no maximum. I tried to enter NA or NULL when there was no minimum or maximum but it didn't work, so the easiest thing to do is to specify 0 for no minimum and the root age for no maximum.
age.min <- c(
0,
280,
270
)
age.max <- c(
354,
354,
354
)
Finally, chronos expects a last column of the data matrix that is not currently used. Still it needs to have the right dimensions:
soft.bounds <- c(
FALSE,
FALSE,
FALSE
)
Now simply unite these four vectors into a single matrix:
mycalibration <- data.frame(node, age.min, age.max, soft.bounds)
The names of the vectors turn into the column names that chronos expects, and off we go: three calibration points. I hope some readers find this useful, and that it works as well for them as it did for me.
Friday, February 26, 2016
Species trees from SNPs with SVD quartets
Today I have tried for the first time to use the SVD quartet method for inferring species trees from Single Nucleotide Polymorphism (SNP) data. This also marks my first foray into the new PAUP test versions.
In case it is unclear what this is about, the situation is that you have several samples per species and genome-wide SNPs for each of them, and you want to infer the species tree, that is a phylogeny that has species instead of your samples as the OTUs, under the assumption of the coalescent model. This post thus ties in with my previous posts on species tree methods for multi-locus data and the Bayesian approach to species trees from SNPs.
And just like those it will not at all be about the theory but about how to do it in practice. For those who fancy dozens of pages of mathematical formulas, the developers of the method, Chifman & Kubatko, have provided the background on arxiv. For the moment, however, I just wanted to try it out.
The newest version of the SVD quartet method is implemented in PAUP, so head over there and download it. I am using Linux, so I have to use the command line version, but that is okay. The main work of setting up a nexus file cannot be avoided anyway.
In case it is unclear what this is about, the situation is that you have several samples per species and genome-wide SNPs for each of them, and you want to infer the species tree, that is a phylogeny that has species instead of your samples as the OTUs, under the assumption of the coalescent model. This post thus ties in with my previous posts on species tree methods for multi-locus data and the Bayesian approach to species trees from SNPs.
And just like those it will not at all be about the theory but about how to do it in practice. For those who fancy dozens of pages of mathematical formulas, the developers of the method, Chifman & Kubatko, have provided the background on arxiv. For the moment, however, I just wanted to try it out.
The newest version of the SVD quartet method is implemented in PAUP, so head over there and download it. I am using Linux, so I have to use the command line version, but that is okay. The main work of setting up a nexus file cannot be avoided anyway.
Wednesday, September 23, 2015
The Templeton Test in parsimony analysis: part 2, how to do it in PAUP and TNT
After discussing the Templeton Test in an earlier post, this post is about how to conduct it in practice.
Saturday, May 30, 2015
Non-hierarchical clustering of mixed data in R
I had wondered for some time how one could do an analysis similar to STRUCTURE on morphological data. Of course not quite like STRUCTURE, as in using a model of population genetics, but in the sense of having a method that gives you the best phenological clusters of your specimens for a given number k of clusters.
The problem was always that I didn't know where to get a method that can handle all data types. You may have numerical variables such as leaf length, where it appears obvious that 5 cm is twice as far away from3 1 cm as from 1 3 cm (ye gods, did I really get that wrong?); but in the same dataset you may also have categorical variables such as hairy or not hairy. These could be coded as 0 and 1 for Euclidean distances, but what if they are multi-state? What is more, you can have categorical variables where there is a clear ordered relationship, e.g. subglobose is somewhere between globose and hemispherical, and others where there is no clear relationships, e.g. white, yellow and purple flower colour.
The R package homals can do a homogeneity analysis, a generalised form of PCA-type ordination analysis, with such mixed datasets. But that is it, it will give you a 2d or 3d plot. It will not do clustering.
This week some concerted googling finally produced the solution: The R package cluster (unsurprising, I know) and its functions daisy and pam (of which I had not heard before). The name doesn't have anything to do with the Asteraceae family, the distance functions in cluster rather appear to be named after female names. When presented with mixed data, daisy defaults to the Bower metric as a distance measure, which is exactly what I wanted.
Because others might be interested in doing non-hierarchical clustering with morphological data, and because getting this right was quite a struggle (as so often with R), I thought I would post a how-to.
The problem was always that I didn't know where to get a method that can handle all data types. You may have numerical variables such as leaf length, where it appears obvious that 5 cm is twice as far away from
The R package homals can do a homogeneity analysis, a generalised form of PCA-type ordination analysis, with such mixed datasets. But that is it, it will give you a 2d or 3d plot. It will not do clustering.
This week some concerted googling finally produced the solution: The R package cluster (unsurprising, I know) and its functions daisy and pam (of which I had not heard before). The name doesn't have anything to do with the Asteraceae family, the distance functions in cluster rather appear to be named after female names. When presented with mixed data, daisy defaults to the Bower metric as a distance measure, which is exactly what I wanted.
Because others might be interested in doing non-hierarchical clustering with morphological data, and because getting this right was quite a struggle (as so often with R), I thought I would post a how-to.
Monday, June 10, 2013
Update on species tree methods
I have updated my earlier post on species tree methods to include three methods made available on a very useful server and to reflect what I have recently learned about PhyloNet, STEM-hy and the speed (or rather, lack thereof) of Mesquite when trying to find MDC species trees for larger datasets. I have also promoted the post to appear among the links on the right.
Monday, May 6, 2013
What to avoid when writing the resume for a job application in science
Not enough time to continue the series on species at the moment. But apropos of nothing in particular, it might be helpful for somebody who searches the web for advice on job applications in science to read what irritates me when going through resumes. Just my own opinion, of course, but it can reasonably be assumed that I am not alone.
So when somebody writes their application for a job as a scientist and compose their CV or resume, they may want to consider the following:
Do not mix your publication list with conference posters or talks. A poster at a conference is not a peer-reviewed journal publication. At best, the recipient of your application will resent having to figure out what is what, at worst they will assume that you are trying to inflate your perceived publication record by deliberately confusing the reader. Keep the two in separate sections.
Do not include submitted manuscripts or manuscripts in preparation as part of your publication list. The word publication kind of implies that the manuscript in question has actually, you know, been published. If it is still in preparation, it may never be. One can have six papers in preparation and then never finish three of them because there is just no time for it at the new job or now that the baby is born, and even submitted manuscripts may be rejected by three journals in a row and then the authors give up on them. That happens. The same applies as in the above item: Make separate lists titled "publications", "submitted manuscripts" and "manuscripts in preparation".
List your publications in a logical sequence, preferably from youngest to oldest, alternatively the other way round; "in press" obviously counts as youngest. Jumbling them up randomly in a non-chronological order makes it harder to see what you have done the last few years and will only annoy the reader.
If you do not quite fulfill one of the criteria or do not have one of the skills for the job, the best strategy is to honestly admit it. It may not be a fatal weakness of your application because there is a chance that no other candidate has all the skills that would make them perfect either. It is not a good strategy to write nothing whatsoever on the topic. Instead you should try to make the case that it would be easy for you to obtain those skills because you are good at learning, that you always wanted to do it anyway but never had the resources to try it, that it is sufficiently similar to something else you have done, something like that. The worst strategy is to write a long and rambling but completely unrelated text block that patently tries to confuse the reader and distract from the fact that you do not have that skill, especially by deliberately misunderstanding the question. People will see through it and they will not necessarily interpret it in your favor.
Apart from that, a reader who found this helpful might also want to check out my old post on job applications in three different scientific communities.
So when somebody writes their application for a job as a scientist and compose their CV or resume, they may want to consider the following:
Do not mix your publication list with conference posters or talks. A poster at a conference is not a peer-reviewed journal publication. At best, the recipient of your application will resent having to figure out what is what, at worst they will assume that you are trying to inflate your perceived publication record by deliberately confusing the reader. Keep the two in separate sections.
Do not include submitted manuscripts or manuscripts in preparation as part of your publication list. The word publication kind of implies that the manuscript in question has actually, you know, been published. If it is still in preparation, it may never be. One can have six papers in preparation and then never finish three of them because there is just no time for it at the new job or now that the baby is born, and even submitted manuscripts may be rejected by three journals in a row and then the authors give up on them. That happens. The same applies as in the above item: Make separate lists titled "publications", "submitted manuscripts" and "manuscripts in preparation".
List your publications in a logical sequence, preferably from youngest to oldest, alternatively the other way round; "in press" obviously counts as youngest. Jumbling them up randomly in a non-chronological order makes it harder to see what you have done the last few years and will only annoy the reader.
If you do not quite fulfill one of the criteria or do not have one of the skills for the job, the best strategy is to honestly admit it. It may not be a fatal weakness of your application because there is a chance that no other candidate has all the skills that would make them perfect either. It is not a good strategy to write nothing whatsoever on the topic. Instead you should try to make the case that it would be easy for you to obtain those skills because you are good at learning, that you always wanted to do it anyway but never had the resources to try it, that it is sufficiently similar to something else you have done, something like that. The worst strategy is to write a long and rambling but completely unrelated text block that patently tries to confuse the reader and distract from the fact that you do not have that skill, especially by deliberately misunderstanding the question. People will see through it and they will not necessarily interpret it in your favor.
Apart from that, a reader who found this helpful might also want to check out my old post on job applications in three different scientific communities.
Wednesday, April 24, 2013
Comparison of species tree methods
Update 10 June 2013: This post originally from 24 April 2013 has been updated extensively because I have since tried out a set of new species tree methods, got STEM to run and a bit more experience with some others. I have also promoted the post to one of the "recommended phylogenetic systematics" posts on this site despite not being about theory of classification.
Update 27 March 2016: Added ASTRAL and iGTP, restructured the post to be more software-focused.
-----
I spent part of the last few days trying out different species tree methods, partly to help a colleague produce an example tree that he can use in a workshop he is planning and partly because I want to infer a species tree for one of my own projects in the next few weeks. This post was written for two reasons: as a note to myself for future reference and as a pointer for somebody who might want to infer a species tree and does not know which of the many programs to choose. A person like myself a few days ago, one could say, only if they find this post via search engine it might save them some of the frustrations I experienced.
Note that this is not a post for a methods wonk or for somebody who wants to learn about the theoretical or methodological background. It is strictly from the end user perspective, directed at those who want to know what is available, how user friendly the tools are and where to get them.
If you don't know what this is about you might want to refer to my earlier post on the topic. To summarize: these days we mostly use molecular data, in particular the DNA sequences of genes or intergenic spacers, to infer the evolutionary relationships of species. However, any individual gene phylogeny may or may not be congruent with the species phylogeny or with other gene phylogenies because each species inherits a random subset of the pre-existing allele diversity in of its ancestral species. Alternatively, discrepancies between a gene tree and the species tree or between different gene trees may also arise from introgression, rare gene flow between distinct species.
Update 27 March 2016: Added ASTRAL and iGTP, restructured the post to be more software-focused.
-----
I spent part of the last few days trying out different species tree methods, partly to help a colleague produce an example tree that he can use in a workshop he is planning and partly because I want to infer a species tree for one of my own projects in the next few weeks. This post was written for two reasons: as a note to myself for future reference and as a pointer for somebody who might want to infer a species tree and does not know which of the many programs to choose. A person like myself a few days ago, one could say, only if they find this post via search engine it might save them some of the frustrations I experienced.
Note that this is not a post for a methods wonk or for somebody who wants to learn about the theoretical or methodological background. It is strictly from the end user perspective, directed at those who want to know what is available, how user friendly the tools are and where to get them.
If you don't know what this is about you might want to refer to my earlier post on the topic. To summarize: these days we mostly use molecular data, in particular the DNA sequences of genes or intergenic spacers, to infer the evolutionary relationships of species. However, any individual gene phylogeny may or may not be congruent with the species phylogeny or with other gene phylogenies because each species inherits a random subset of the pre-existing allele diversity in of its ancestral species. Alternatively, discrepancies between a gene tree and the species tree or between different gene trees may also arise from introgression, rare gene flow between distinct species.
Wednesday, February 13, 2013
How to give a good scientific talk
Recently I got drawn into a short discussion on how best to give scientific talks. As a reaction to that discussion, the following is in part a note to myself and in part a piece of advice in case an early career scientist happens on this blog via a search engine and finds this useful. As always, while the following is based on my own experience as a career scientist speaking at various conferences from ca. 25 to more than 2,000 participants, please note that I am not a professional instructor for public speaking courses. In addition, the culture of speaking will vary from one area of research to the next, and what is appropriate in mine may or may not be appropriate in yours.
What we are talking about here are the typical self-submitted, non-keynote talks at scientific conferences. They generally last 12 min plus 3 min for questions or 15 min plus 5 min for questions, and the resources at your disposal are generally PowerPoint slides projected at the wall behind you, a laser pointer and a microphone. You may have a remote to advance the slides or you stand behind a lectern with a keyboard and press the arrow keys to change slides. So here is my advice.
What we are talking about here are the typical self-submitted, non-keynote talks at scientific conferences. They generally last 12 min plus 3 min for questions or 15 min plus 5 min for questions, and the resources at your disposal are generally PowerPoint slides projected at the wall behind you, a laser pointer and a microphone. You may have a remote to advance the slides or you stand behind a lectern with a keyboard and press the arrow keys to change slides. So here is my advice.
Monday, December 17, 2012
Readability scores
As a scientist, I am writing a lot, especially manuscripts and grant proposals. Next to content and structure, one of the aspects that has the greatest impact on whether somebody else will appreciate or even be able to make sense of what we write is the complexity of the language we use. Long and convoluted sentences are off-putting and confusing, and the use of unnecessarily technical terms can come across as pretentious. In the worst case, we may write a text that most of the intended audience cannot understand, especially if we are a specialist addressing non-specialists.
Now obviously we can have a hunch about this, but if in doubt we can use various metrics that have been developed to provide quantitative tests of readability: the automated readability index, the Flesch reading ease score, the Flesch-Kincaid grade level, the Coleman-Liau index, the Gunning fog index and the SMOG index.
Now obviously we can have a hunch about this, but if in doubt we can use various metrics that have been developed to provide quantitative tests of readability: the automated readability index, the Flesch reading ease score, the Flesch-Kincaid grade level, the Coleman-Liau index, the Gunning fog index and the SMOG index.
Tuesday, December 4, 2012
The Google alternatives to ResearcherID and Journal Citation Reports
In a previous post, I explained how to use ResearcherID. In my post on assessing publication records, I explained what an h-index is and mentioned how many people judge journals by their impact factors. ResearcherID, which you can use to automatically calculate your h-index and other metrics, and the Journal Citation Reports (JCR) are run by Thompson Reuters, the first as a free service, the latter as a subscription service. However, Google has recently introduced its own variants of these services, and both are free. This post examines how they differ from the Thompson Reuters products and explores their advantages and disadvantages.
Sunday, December 2, 2012
How to use ResearcherID
ResearcherID is a free service offered by Thomson Reuters that is very useful to scientists for two reasons: First, it allows you to select all your publications and connect them to the eponymous ID. This is especially important if you have a relatively common name, because in that case some potential employer who wants to look you up in a publication database will find a confusing number of entries from many different people. With ResearcherID, you can put the ID onto your website or CV, and people searching for your ID will find your and only your publication record.
Second, you may need to calculate your citation metrics - most frequently lifetime citations and h-index - for an internal report or for the CV that goes with a job or grant application. It is annoying to have to calculate these metrics from scratch every time you need them. With ResearcherID, you merely have to tell the system once which publications are yours and then add any new publication when it appears. If you keep the list updated in that way, ResearcherID will automatically calculate these and other metrics every time you look up your own profile (simply click on "Citation Metrics" under "My Publications" on the left and wait a few seconds).
Second, you may need to calculate your citation metrics - most frequently lifetime citations and h-index - for an internal report or for the CV that goes with a job or grant application. It is annoying to have to calculate these metrics from scratch every time you need them. With ResearcherID, you merely have to tell the system once which publications are yours and then add any new publication when it appears. If you keep the list updated in that way, ResearcherID will automatically calculate these and other metrics every time you look up your own profile (simply click on "Citation Metrics" under "My Publications" on the left and wait a few seconds).
Friday, November 30, 2012
How to assess publication records
Publish or perish has become such a well-known expression that it should be nothing new even to those who have never played with the thought of pursuing an academic career (see Wikipedia, or, more cynically, SMBC). The idea is that there is ever increasing pressure on academics not necessarily to produce work of high utility and quality but simply to produce lots of publications in renowned journals.
Academics are evaluated on the basis of their publication record in nearly all professionally relevant situations: when applying for a job, when considered for a promotion, when applying for a research grant. By extension, research groups within an institute, institutes within a university, and universities among universities are evaluated based on the publication record of their researchers. So, how does one assess publication records?
There are different ways people do this, and they are listed below in order of decreasing stupidity.
Academics are evaluated on the basis of their publication record in nearly all professionally relevant situations: when applying for a job, when considered for a promotion, when applying for a research grant. By extension, research groups within an institute, institutes within a university, and universities among universities are evaluated based on the publication record of their researchers. So, how does one assess publication records?
There are different ways people do this, and they are listed below in order of decreasing stupidity.
Subscribe to:
Posts (Atom)




















